Abstract
Machine learning algorithms have shown great accuracy in predicting chemical reaction outcomes and retrosyntheses. However, designing synthesis pathways remains challenging for existing machine learning models which are trained for single-step prediction. In this manuscript, we propose to recast the retrosynthesis problem as a string optimization problem in a data-driven fingerprint space, leveraging the similarity between chemical reactions and embedding vectors. Based on this premise, multi-step complex synthesis can be conceptualized as sequences that link multidimensional vectors (fingerprints) representing individual chemical reaction steps. We extracted an extensive corpus of chemical synthesis from patents and converted them into multidimensional strings. While optimizing the retrosynthetic path, we use the Euclidean metric to minimize the distance between the expanded trajectory of the growing retrosynthesis string and the corpus of extracted strings. By doing so, we promote the assembly of synthetic pathways that, in the chemical reaction space, will be more similar to existing retrosyntheses, thereby inheriting the strategic guidelines designed by human experts. We integrated this approach into the RXN platform (https://rxn.res.ibm.com/) and present the method’s application to complex synthesis as well as its ability to produce better synthetic strategies than current methodologies.
Similar content being viewed by others
Introduction
The retrosynthesis is the process of designing a synthetic route for a desired target molecule and requires the identification of optimal strategies to combine simpler molecules into a target product1. Frequently, retrosynthesis entails a series of reaction steps to synthesize those molecules from simpler precursor molecules. One of the main challenges in this process is exploring the large retrosynthesis hypergraph, which represents all possible synthetic routes for a given target molecule2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30. The pathways within the tree link the target product (i.e., the root) with all commercially available compounds (i.e., the leaves) which are identified by the algorithm through single-step disconnections.
The retrosynthesis tree is exponentially large because each retrosynthetic step can potentially branch into multiple alternatives, and the number of possible routes increases exponentially with the depth of the tree. Consequently, the sheer volume of potential synthetic routes can be overwhelming to explore using classical mathematical kernels, even for relatively small molecules.
The exploration of such hypergraphs requires the implementation of specific criteria to effectively filter the extensive array of disconnection options. One strategy relies on the evaluation and scoring on the pathways based on the confidence of each single-step retrosynthesis prediction, which can be individually evaluated. The idea is to consider the low confidence as a metric for an highly risky and most likely failing synthetic route. Therefore, steps with low confidence are not further propagated, giving higher priority to those with higher confidence14. Apart from filtering based on confidence values, single-step predictions that fail the round-trip check can also serve as additional targets to avoid further expansion. This check consists of applying a forward prediction model to the output of the single retro-step prediction and of verifying whether or not the result of this operation returns the desired product14. In other schemes, one could score and rank the different options, for example depending on the molecule availability, the corresponding cost, or by some metric related to green chemistry31.
Notwithstanding, most of the existing approaches exclusively use local information derived from single-step retrosynthesis and do not keep into account strategical decisions typical of a multi-step synthesis conceived by a human expert. In fact, when undertaking a multi-step synthesis, various strategic decisions can be made to streamline the process and optimize efficiency. One example is the strategic protection of functional groups. By selectively safeguarding specific groups early on, chemists can prevent unwanted reactions and ensure the desired transformations occur smoothly. In ideal cases, chemists follow the idea of introducing different protecting groups, targeting the removal of all protections in a single step at the final stage, saving time and minimizing the risk of side reactions. Another strategy involves utilizing robust and high-yielding reactions for key transformations, which can significantly impact the overall yield and simplify the synthesis route. Additionally, strategic retrosynthetic disconnections play a crucial role in planning the sequence of reactions. By identifying strategic bond disconnections, chemists can design efficient synthetic pathways and target specific intermediates or building blocks to assemble the final product. Lastly, the choice of reagents, catalysts, and reaction conditions is another strategic consideration. Selecting appropriate reaction parameters can enhance selectivity, improve yields, and expedite the overall synthesis process. These strategic decisions collectively contribute to the successful execution of complex multi-step syntheses32. Due to their dependence on single-step predictions, existing models lack a comprehensive understanding of the key strategies employed in multi-step retrosynthesis.
Recently, Thakkar et al.29 described an approach aiming to improve retrosynthetic prediction systems by allowing chemists to have more control over the disconnections made during the exploration of the retrosynthesis tree. The method enables user-defined disconnections, creating a “human-in-the-loop” component that combines expert knowledge with deep learning. Their approach results in an increased diversity of predicted disconnections. With their method they improved decision-making strategies thus enhancing the chemist’s experience and facilitating user engagement that statistical and machine learning algorithms alone cannot encode due to insufficient training data and resulting model biases. Another recent approach to improve the search policy builds on reinforcement learning to introduce a notion of goal-driven synthesis planning that optimizes multistep synthesis routes toward specific building blocks33.
Furthermore, Chen et al.21 introduced a method called “Retro*”, using an innovative neural-guided tree search approach for chemical retrosynthesis planning. Their method uses an A-like planning algorithm guided by a neural network trained on past retrosynthesis planning experiences. Their neural network learns synthesis costs for each molecule, assisting the search algorithm in choosing the most promising molecule node for expansion. In addition, the work by Ishida et al. introduces “ReTReK”, a data-driven computer-aided synthesis planning (CASP) application that integrates retrosynthesis knowledge into the evaluation of search directions. By incorporating adjustable parameters based on retrosynthesis knowledge, ReTReK successfully explores promising synthetic routes, demonstrating its preference for routes designed with the knowledge. The study addresses limitations in existing data-driven CASP applications by introducing rule-based techniques and evaluating performance using drug-like molecules, showcasing ReTReK’s potential to enhance both current and future data-driven CASP applications.
Recently, Pasquini and Stenta30 showcased LinChemIn, a toolkit that simplifies the manipulation of reaction networks and enhances functionality for working with synthetic routes, promoting interaction between AI and human expertise in chemical analysis.
For a comprehensive review, we refer the reader to the evaluations provided by Zhong et al.34 and Jiang et al.35.
However, the problem of designing chemical retrosynthesis is far more complex than removing potential biases from single-step retrosynthesis model or to steer routes toward specific precursors. It involves knowledge, experience and also a certain degree of creativity and intuition which goes beyond the state of the art of existing retrosynthesis algorithms. Similar to a strategy game, the evaluation of multi-step solutions requires holistic planning and thus may be more effectively conducted by considering a sequence of steps rather than solely focusing on individual steps. Hence, relying exclusively on the confidence of a single-step model to devise a synthetic route can potentially overlook crucial pathways and lead to sub-optimal or even erroneous predictions.
Our algorithm not only addresses strategic decisions but also extends its impact to enhance the efficiency of separation steps in multi-step synthesis planning. Unlike traditional approaches that rely solely on single-step retrosynthesis models, our method introduces a strategy for assembling single-step predictions into coherent retrosynthetic pathways.
By considering the entire sequence of steps, our approach provides a broader view of the synthesis process, including the crucial final stages where separation efficiency is crucial.
In this study, we present an algorithm that emulates human strategic decision-making when constructing an AI-driven retrosynthesis approach. The computational method facilitates the exploration of the retrosynthesis tree, which is constructed using conventional single-step machine learning predictions, leveraging the chemical knowledge derived from existing collections of multi-step retrosynthesis. By doing so, the algorithm effectively harnesses the expertise and available knowledge of human chemists readily accessible through retrosyntheses published in literature. The proposed method targets the task of assembling efficiently the single-step retro predictions and does not require any retraining of retrosynthesis models, since it makes used of existing pre-trained models. The algorithm focuses on using an embedding to represent sequences of chemical steps. It then compares the sequences of predictions with the sequences of steps in pre-existing datasets to prioritize retrosynthesis strategies. For the representation of the single-step chemical reaction, we leverage the work of Schwaller et al.36, who utilized embeddings of language models to build a chemical reaction fingerprint (rxnfp). Such reaction fingerprints capture structural and chemical properties, such as reactants, products, reaction context, and stereochemistry. These embeddings proved to be very successful to relate chemical reactions to specific reaction classes37, to predict reaction yield38 or even to discovery novel Heck reactions39.
Here, we extend the concept of chemical reaction fingerprints to retrosynthesis routes, representing the sequences of steps contained in published retrosynthesis with a set of multidimensional strings in the fingerprint space. The core idea of the proposed algorithm is to construct the retrosynthesis tree by growing strings minimizing the distance between the predicted and each section of any pre-existing multidimensional string in the embedding space. The comparison can be extended to more intricate scenarios, where retrosyntheses are not linear trajectories but rather trees depicted by corresponding branched structures in the fingerprint space.
This method demonstrates better performance in terms of producing retrosynthesis with a smaller number of steps, protects/deprotects functional groups making decision across the entire length of the synthesis. The proposed approach makes better use of the different available reactions leading to steps which can occur in milder reaction conditions, avoiding for example the need for strong chemicals like organometals and powerful oxidants. We provide a few applications in the results section, which are illustrative of the potential of the methodology.
Results
Data preprocessing and preparation
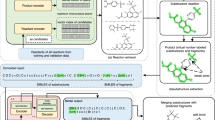
Our method relies on representing retrosynthesis routes as trajectories in the fingerprint space as described in the “Method” section. We assembled the corpus of retrosynthesis routes by processing the Pistachio dataset40, which is a large-scale dataset of chemical reactions extracted from US patents. The dataset contains over 4 million reactions. We assembled 1,202,092 linear and 187,478 branched pathways. Figure 1a illustrates the distribution of number of steps for the linear and branched pathways consisting of at least two steps.
To maximize the extraction of the human expert knowledge contained in all sequences, we extracted from each retrosynthesis route additional sub-routes of varying lengths, from a minimum of two steps up to the maximum length of each pathway. For example to build the datasets of sequences N-step long, we utilize all the retrosynthesis with length greater or equal to N. A retrosynthesis of length M, with M greater then or equal to N, produces M + 1 − N sequences of length N. However, if the number of steps exceeds 15, we truncate the sequence at 15 steps because, we explained in the “Method” section, the use of longer sequences does not add any performance improvements. Figure 1b shows the final distribution of lengths of all sequences in our dataset. Figure 2 shows the Euclidean distance distribution of the fingerprints of the reaction SMILES in the Pistachio dataset. The histogram is the distribution of distances between randomly selected pairs of chemical reaction fingerprints. The number of pairs is large enough to give a converged distribution, which provides a numerical information on the range-distance values in the fingerprint space. We also observe a peak at shorter distances, centered at about 6, which relates almost entirely to distances among reactions from the same class, see Fig. S1a in the Supplementary Material. We verify this finding by computing the distance distributions of groups of chemical reaction SMILES belonging to the same reaction class, which we report in Supplementary Fig. 1b. In addition, we computed the distance distribution of chemical reaction SMILES in the same class and in different classes using the curated dataset by Schneider et al.41. Supplementary Fig. 1c shows a two-peaks distribution with a clear separation between the distribution of distances of SMILES in the same class, first peak, compared to the distribution among the remaining SMILES. Compared to Supplementary Fig. 1a, Supplementary Fig. 1c shows a more distinct separation between the reaction records compared to a more complex dataset. This clearer delineation can be attributed to the reduced number of reaction types in the simplified dataset of Schneider et al.41. We repeated the same analysis using cosine distance instead of Euclidean, which showed the same two-peak distribution. In the rest of the work, we will use the Euclidean distance because it better resolves the distribution of fingerprints in the same class compared to other metrics. To accelerate the computation and memory consumption, we employed PCA reduction, reducing the fingerprint embedding from 256 to 16 dimensions without significant loss of descriptive capacity, see “Methods” section for details. Figure 2 shows the distribution of distances in the reduced space and compared it with the original space. Our observations reveal that the PCA dimensionality reduction improves the separation between short-range and long-range distance peaks in both Pistachio and Schneider datasets.
The Euclidean distances have been computed in the original fingerprint space of 256-dimensions (solid lines) and compared to the PCA 16-dim reduced space (dashed lines). The frequency of chemical reaction SMILES within the same class were centered around values of 6 in the original fingerprint space. Both plots have been normalized by the same total number of pair counts. The Pistachio distribution has been computed on subset of Pistachio data. In this case, the number of data points is large enough to give a converged distributions.
Method evaluation and comparative analysis
To evaluate the performance of our approach, we performed a comparison of this method based on multidimensional strings (see “Method” section) with an hypergraph search strategy14 on a series of case studies.
We analyzed the routes obtained with both approaches on two molecules that are challenging for a search strategy based on hypergraph exploration (Product A and B in Fig. 3).
Our results demonstrate several advantages of our string-growth method compared to alternative approaches that rely on single-step confidences. Figures 4 and 5 show for both molecules the predictions obtained with the two approaches. In Fig. 4, the proposed method finds solutions with less steps. With the standard approach the retrosynthetic tree counts 8 reaction steps, compared with 5 steps of the proposed method. Using the same settings and the same molecule, the proposed algorithm reaches commercial precursors more easily. Analyzing more carefully each step, it is possible to observe that the first two reactions are mostly the same: Amide formation at the first step and protection of the phenol at the second step, although the protecting group is different. From the third step instead the disconnections are different: with our algorithm, the intermediate is obtained through Heck coupling, without any possible interference, and the relative precursors are obtained in a single step from commercially available compounds. On the contrary, using the older algorithm the double bond is made through Horner-Wittig reaction, reaction that involves the use of strong bases, after the mesylation of the phenol, that due to his acidity can interfere. Even the two precursors of the Horner-Wittig reaction are not commercial and have to be made through complex reactions. Given these observations it seems like the hypergraph approach leads to worse intermediates during the retrosynthesis and it struggles to reach commercially available compounds, so that it has to force it through low confidence chemistry. Instead, in the proposed algorithm, the retrosynthesis is well designed and that is confirmed by the its shortness and higher reliability.
Retrosynthesis paths obtained with the proposed method (right) and with the standard confidence-based approach (left) for Product A of Fig. 3.
Figure 5 illustrates an analogous comparison for Product B. The retrosynthesis depicted in Fig. 5 showcases two comparable routes in terms of length, but they differ in the choice of reactions employed. Both retrosynthetic pathways share the initial step of amide formation via the Steglich reaction, leading to the same set of intermediates. In the string-growth approach, the carboxylic acid is obtained through saponification of a commercially available ester, while the standard route involves the predicted formation of an ether through nucleophilic aromatic substitution. Although both routes are reliable, the saponification process is considerably easier to execute and thus preferable for chemists. In terms of the amine, both routes involve deprotection of a Boc protecting group. With the hypergraph exploration method, the bicyclic structure is made using a Simmons-Smith reaction from the alkene and the last disconnection is a Suzuki-Miyaura coupling. In the proposed approach the aromatic ring is obtained through [3 + 2] cycloaddition, favored by the aromaticity of the product, preceded by the formation of the oxime. The two path are built through different choices: in one case it is chosen to form the bicyclic structure and on the other side the formation of the isoxazole. Also in this case it has to be highlighted the preference of our approach for a more direct chemistry rather than reactions conducted under harsh conditions, despite the more direct path chosen.
Retrosynthesis paths obtained with the standard confidence-based approach (left) and with the proposed method (right) for the Product B of Fig. 3.
In the Supplementary Material, we show the retrosynthesis paths of Product A and B obtained via the method proposed by Chen et al.21, see Supplementary Figs. 5 and 6, respectively. In these examples, we observe that the reaction pathways generated via the Retro* method are sub-optimal due to non-regioselective reactions.
To further analyze the impact of the proposed search strategy compared to a single-step confidence-based, we considered four additional examples where the latter is performing well. We employ the same settings as in the previous case studies with a focus on delineating the distinctions between the an hypergraph exploration approach and the proposed method. First, the comparison reported in Fig. 6 shows how predicted reaction pathways can exhibit a lot of similarities. The key distinction lies in the formation of the heteroaromatic chloride. The standard confidence-based approach relies on it being present in the building block, whereas the proposed method involves its creation under harsh conditions with POCl3. Nevertheless, this step streamlines the retrosynthetic process, allowing to initiate synthesis from a renewable building block.
Second, in Fig. 7 we can observe how two predicted routes can diverge in the construction of the core of the target molecule. The proposed method initiates with a cycloaddition to shape the three-membered cycle, and safeguarding the hydroxylic group enables manipulation of the opposing segment. In contrast, the standard confidence-based approach uses a Wittig reaction, followed by a hydroboration-oxidation sequence to achieve a comparable outcome. Third, in the molecule considered in Fig. 8, we observe how the initial two disconnections remain identical. Subsequently, the proposed method capitalizes on the selectivity arising from two distinct halogen atoms. Ultimately, it tries to obtain the bicyclic aromatic structure with a nucleophilic substitution-carbonylamine condensation. Lastly, in Fig. 9, the solution derived from the proposed method is more concise as it directly commences from the spyro-compound. In contrast, the hypergraph exploration method expends considerable effort in its construction. This underscores the string-growth approach’s proficiency in identifying a more direct route from a commercially available precursor. Additionally, the retrosyntheses are equivalent, differing only in the choice of protecting groups for the aminic group and the methodologies employed for its formation.
The overall effect of the proposed search approach is that the steps along the retrosynthetic tree are more interconnected, less focused on a single reaction and more on the quality of the resulting sequence, leading to shorter retrosynthesis with an higher quality and reliability of each step.
Figure 10 and Fig. S4 in the Supplementary Material compare the length of the retrosynthesis obtained with the hypergraph search and our proposed method, which produces on average less deep retrosynthesis trees. The retrosyntheses planned by our proposed method overall consist of fewer reactions, see Fig. S7 in the Supplementary Material, which compares the total number of reactions of retrosynthesis predicted via hypergraph search, our proposed method, and the “Retro*” method21.
The histogram focuses on the frequency of tree depth for each product based on the top-1 results. Vertical blue and orange dashed lines indicate the means of the two distributions: hypergraph search and the proposed method, respectively. In the Supplementary Material Fig. S4 provides additional comparisons of the length of predicted retrosynthesis.
For reproducibility, in the Supplementary Material, we report the parameters configuration used in the RXN for Chemistry platform to produce the reported results, see Supplementary Fig. 2.
Discussion
We present a novel method for generating complete retrosynthesis pathways that incorporates human expert synthesis strategies, which are not captured by existing single-step prediction models. Our method utilizes chemical reaction fingerprints, commonly used for reaction classification, to capture multi-step strategies. Retrosynthetic routes are represented as strings in the fingerprint space. The fingerprints of published chemical routes are used to populate a database, which is then used to guide the expansion of the retrosynthesis tree by ranking branches using a score that favors pathways closer to those compiled by human experts. This approach prioritizes pathways that use similar retrosynthetic strategies, without requiring additional training of existing single-step prediction models.
Because at the time of this publication there are no benchmarks available for multi-steps synthesis, the testing a scale poses significant challenges. Therefore, we have engaged expert chemists among our authors to evaluate specific examples.
We demonstrate the effectiveness of our method by applying it to the synthesis of several products, which serve as prototypical examples to highlight the differences between our approach and the older statistical approach. In general we observe that the proposed algorithm shows improved results by creating shorter retrosynthesis paths. It efficiently handles the protection and deprotection of functional groups throughout the entire synthesis process. The method maximizes the utilization of various reactions, allowing for gentler reaction conditions for example eliminating the requirement for harsh substances such as organometals and potent oxidants.
Our proposed algorithm, which currently rewards synthesis routes closely resembling those documented in the Pistachio dataset, is designed to be adaptable to various preferences and constraints within the fingerprint space. By default, our scoring metric considers all the retrosyntheses in Pistachio equally important. Nothing prevents to customize our score function by assigning different weights to routes based on specific preferences or constraints, or, alternatively, combining with other metrics linked to trajectories in the fingerprint space, for instance, accounting for precise and unambiguous comparisons for improved resource management proposed by Andraos42 or including concepts such as the one of strategic molecules proposed by Weber et al.43.
Methods
Our method relies on representing single-step chemical reactions with reaction fingerprints36. We build sequences of reaction steps of different lengths from the protocol synthesis contained in the Pistachio dataset40 belonging to the same document ID. Each sequence of steps will be referenced as a string or trajectory in the fingerprints space. We compute the fingerprint of a synthetic pathway by concatenating the fingerprints of each step of the sequence and thus construct a database for all the synthetic procedures extracted from Pistachio. To promote the exploration and continuation of branches in the retrosynthesis tree that closely align with pre-existing trajectories derived from publications, we compare distances between the expanded pathway and any pre-existing string. The pathway with shorter distances to existing pathways will be prioritized for further exploration.
Chemical reaction fingerprints
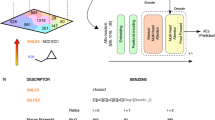
Each step is identified by a chemical reaction in SMILES notation, which consists of a text string containing the precursors and the corresponding product separated by the “>>” token. The precursors are separated by a “.” token. In this work we used the fingerprints of Schwaller et al.36, which is an array of real numbers of length 256. The fingerprints of chemical reaction SMILES of the same reaction class are closer in space compared to the SMILES of different reaction classes.
Dimensionality reduction of the fingerprints of a chemical reaction
Expanding a retrosynthetic route close to humanly compiled pathways requires the operation on fingerprints of thousands of sequences during a single retrosynthesis prediction task. Each operation involved the calculation of the Euclidean distance between pairs of fingerprints. To speed up the computation, we performed a PCA reduction on the set of fingerprints of the chemical reactions, reducing the dimensionality from 256 to 64, 32, 16, and 8. We then computed the correlation between the distances in the original space and the reduced space and we identified a value of 16 being big enough to preserve the relative distances among chemical reaction SMILES.
Retrosynthesis route fingerprints
Given that each single reaction step is a point in the fingerprint space, a sequence of steps is can be thought of a string connecting the individual steps part of the synthetic pathway. Figure 11 shows two examples: for a linear and branched synthesis, see Fig. 11a, b, respectively. The string contains the information both on the order of the steps in the sequence, but also on their nature. Branched synthesis, similar to linear synthesis, can be described in the same space, as it is shown in the second example illustrated in Fig. 11b.
a Illustrative representation projected in two dimensions of sequence of chemical steps in the fingerprint space. This sequence could represent an entire linear synthesis or a smaller part consisting of six consecutive steps. Each of the six chemical steps maps to a specific point in the embedding space. The sequence is represented by a trajectory connecting all the points one after the other with a specific order. For visualization purposes, a two dimensions fingerprint space is used. b Same illustrative example as in (a) but for a non-linear synthesis, described via tree structure. Different trajectories merge in common nodes. c Graph representation of the retrosynthesis tree illustrated in (b). d Linear sequences derived from retrosynthesis tree. Each sequence connects one leaf of the tree to the common root. The root of the tree represents the last (time ordered) reaction step giving the target product. The root in at depth equal to zero. The leaves are at depth greater than zero. We use the convention to count the steps from the root to the leaves (backward).
The fingerprint of a retrosynthesis route (or string) is represented via an array built concatenating the fingerprint of each chemical reaction step, in the same order they appear in the sequence. A sequence of length N would be described by an array of N × 256 dimensions, if we use the original embedding space. The same sequence could be analogously described by an array N × 16 long using the representation in the PCA reduced space. This definition makes every string’s fingerprint depend on its particular length.
For example, considering the case of a linear pathway consisting of M = 6 steps as reported in Fig. 11a, we can build M + 1 − N fingerprints of length N (N ≤ M). It is immediate to see how each retrosynthesis route will contribute only to sets shorter or at most of length equal to the maximum length of the extracted route. Similarly, we can extract linear sequences from branched pathways to augment the sequence dataset. An example is described in Fig. 11.
To expedite the search of closest sequences, we employed the cKDTree class from the scipy library, which allows us to efficiently index each list of vectors and quickly retrieve the nearest neighbors of any given point. This indexing process was performed directly in the PCA reduced space, offering the advantage of a smaller indexed object size that needs to be stored and loaded into memory during runtime.
Algorithm of the fingerprint-driven retrosynthesis
The proposed method requires as input a target product, a single-step prediction model to preform single-step retrosynthesis predictions, and a model to convert chemical reaction SMILES into fingerprints. We use the fingerprint model of Schwaller et al.36 and for the retro-predictions we use the models available on the RXN platform44, with the results presented in the next section relying on the use of the model with ID “2020-07-01”.
In retrosynthesis planning, a route is considered complete when all leaves of the retrosynthesis tree contain reaction SMILES with the precursors that are available. To this end, we use the database provided by eMolecules45 to determine the availability of each molecule. The method involves a series of steps, which are exemplified in Fig. 12 for better understanding. The figure presents a simplified example of a reaction SMILES tree, along with the intermediate levels, highlighting the role of the precursor compounds.
The color code is indicated in the legend of the figure. This representation not only indicates the reaction SMILES, it also shows the composition of the precursors of each SMILES. Green filled dots indicate available compounds, while black filled product are not available precursors that need further steps of synthesis.
Given a target product, the retrosynthesis route is expanded using the following steps:
-
(1)
The first step in our approach is to conduct a top-N single-step retro prediction for the desired product. For each prediction, we generate a reaction SMILES that follows the format of “precursors >> product”, where the precursors are separated by a dot. Subsequently, we calculate the fingerprint of each reaction SMILES.
-
(2)
We filter out all predictions that fail the round-trip check, in which a forward prediction model (in our case the Molecular Transformer46) is used to confirm the correct prediction of the product from the precursors. This check is carried out by using the precursors predicted from the retro model as input for a forward model. If the resulting product from the forward prediction is the same as the original product, we consider the round prediction to be successful.
-
(3)
Our model predicts not only the reactants but also the reagents, solvents, and catalysts, leading to several single-step retro-predictions that may be quite similar. Although they have the same disconnection point, they use slightly different precursors. To ensure better exploration of the retrosynthesis tree and promote diversity, we use the chemical reaction fingerprints to remove similar predictions, that are too close in the fingerprint space. We use a fixed threshold of 2.0, which we determined by examining the distance distribution of these top-N predictions and testing thresholds between 1.0 and 2.5. The top-N predictions, which passed the round-trip check, are sorted by length with its maximum value being N. For each i in [1:N] and each j in [i + 1:N], if dij < 2.0, we remove j. This filter action eliminates reactions that are too similar, expanding further only one of them, while saving computational time as the tree grows deeper. This step will retrain K single-step predictions, with K ∈ [1, N].
Different to our previous approach14, here we do not filter based on the confidence of the forward model, thus avoiding biases on reaction classes that are under-represented and not sufficiently learned. In fact, sometimes there are effective retrosynthesis pathways that involve a single retro-step with low confidence, which would be penalized by confidence-based methods and possibly never explored.
-
(4)
For each of the K results obtained from the previous step, we examine the precursors to identify those that are not readily available. These precursors are deemed intermediate products that require synthesis. We maintain a list of completed reaction SMILES and those that require further prediction steps. Reaction SMILES with all available precursors are marked as completed. Completed pathways including the final product are considered successfully finished retrosynthesis, as the product can be directly obtained from the available precursors. In the example shown in Fig. 12, the reaction labeled s1 is completed in a single step, and the corresponding chemical SMILES (s1) is located at depth zero.
When possible, our approach will return multiple routes, which are compatible with the time and execution constraints provided by the user. In the example of Fig. 12, the prediction s0 contains two available precursors, A and B, and a third precursor that is not available. If compatible with the user settings, the method will continue to search for more routes expanding the leaves of the tree which correspond to reaction SMILES containing one or more non-available precursors. Precursors that are not available are considered intermediate products that need to be synthesized. During the execution we keep updating the lists of chemical reaction SMILES, which contain available precursors, and those that require further prediction steps. For example, in Fig. 12, the reaction labeled s1 is completed in a single step, and the corresponding chemical SMILES is located at depth zero. If the maximum number of prediction is reached, the algorithm returns the results and terminates the execution, otherwise it continues by expanding the tree.
-
(5)
The tree expansions involve single-step retro predictions using as input all the non-available precursors of the non-completed reaction SMILES. In the example shown in Fig. 12, these new predictions are located at depth one and are labeled as s2, s3, and s4.
If the precursors of each of the predictions s2, s3, and s4 were available, the chemical smiles s2, s3, and s4 would be leaves of the tree, and the solution would have three alternatives. In this case, we would return four routes: a single-step route [s1] and three 2-steps routes [s0, s2], [s0, s3], and [s0, s4]. For each route, we would compute the score and rank the solutions accordingly.
However, following the example of Fig. 12, if two precursors of s2 were not available, we would continue the search for more routes involving s2. This process is repeated for each non-completed reaction SMILES until all routes have been explored or the maximum number of solutions requested by the user has been reached.
-
(6)
During the expansion of the retrosynthesis tree, the number of unfinished routes may increase, depending on the number of unavailable precursors. As this number grows, the algorithm computes a score to select which routes have to be further expanded and which should not. The selection is necessary to limit the exploration in favor of the computational time. Continuing with the example of Fig. 12, we now compute the score for pathways consisting of two steps, for example s0 and s2. We compute the fingerprints of s0 and s2, and then we apply the PCA model: each fingerprint after PCA is a point in a 16-dimensional space. After concatenation of the two 16-dimensional arrays into a single array of length 32, we search for the closest five trajectories in the database using the cKDTree library. We store the average distance of the top five closest trajectories as a measure of how close or far the predicted [s0, s2] pathway is from the reported reactions. The same protocol will be applied for computing the score for all other alternative routes, [s0, s3] and [s0, s4] in the example. Although the sequences considered in this description consist of a length of two steps, as the tree depth grows, the score will compare longer sequences.
Among the unfinished routes, we select to expand further only a fraction, typically the top-M having the smallest distance from the database. The number M is usually between 10 and 20. The user can set the value of M to balance between the duration of the prediction and accuracy. Increasing M allows for the continuation of more routes, which results in more exploration but also a larger number of single-step retro predictions and a longer overall prediction time. On the contrary, a smaller value of M reduces exploration for a faster prediction.
After identifying the top M pathways, we search for intermediate compounds that are not available and require further synthesis. These intermediate products are stored in a list, and we perform single-step retro predictions to determine their precursors. In the simplified representation shown in Fig. 12, these predictions are labeled as s5, s6, s7, s8, and s9. However, some predictions may result in invalid SMILES, such as s6, which are discarded.
If the precursors of s5, s8, and s9 are all available compounds, they do not require additional synthesis steps. At depth equal to two, there are three complete routes. One route is given by the single-step reaction s1. The other two routes are given by [s0, s2, s5, s7] and [s0, s2, s4, s8], both of which share the first two steps, [s0, s2].
The algorithm loops over the precursors and labels reactions as finished or to be continued based on the availability of compounds. The system also verifies that all branches of the tree have leaves containing only available compounds to compute the number of completed routes. Once the tree’s status is updated, we verify a set of stopping criteria that the user can set via the user interface.
The algorithm stops when at least one of the following criteria is met. The first termination is effective if the number of completed routes is greater than a user-defined threshold. At each prediction step, which occurs every time the tree increases in depth, there is a check to update the number of the new finished routes. The second termination criteria checks if the maximum depth of the tree is reached. Typically, the maximum depth is set to be between 6 and 15. Finally, if the runtime is exceeding a user provided value, the algorithm will terminate its execution. Whenever a stop condition is met, the execution proceeds directly to the final evaluation of the score and returns the solutions. If none of the stopping criteria are met, the algorithm proceeds to the next step.
-
(7)
As the sequences of steps become longer, further single-step retrosynthesis predictions are required. In this step, we describe how we compute the score for sequences of length greater than two, which was already been presented in the previous step.
To compute the score for a sequence of length N, we consider all sub-sequences of lengths ranging from two to N-1, with a maximum length of 15. We set a threshold of 40 for the distance between two steps in a sequence. This distance is computed in the 16-dimension embedding space after PCA. Any distance greater than the threshold is assumed to be equal to the threshold. This decision is based on the observation that if two steps belong to different classes, their distance is likely to be larger than the first minimum in the distance distribution. The threshold of 40 is already a much larger value than the location of the first minimum. Sub-sequences longer than 15 steps are excluded since it is very improbable to find such long sequences of steps close to each other. For each of these sequences, we search the corresponding database for the closest fingerprints and take the average distance of the closest five as the partial score. We compute the overall score of a sequence as the sum of partial scores from all sequences of different lengths within a specified range, thus accounting for different alignments between predicted sequences and those in the database.
The different pathways are ranked based on their scores, and those with the lowest scores are further expanded in the next single-step prediction iteration. For each expanded pathway, we search for all the nodes in the tree that are necessary to complete the retrosynthesis, which in turn requires all the leaves to represent available compounds. These additional nodes are expanded independently of their score.
-
(8)
After the stopping criteria is met, we compute the score of each route. The route can be either linear or a tree. In the case of a linear route, the score is calculated as described in the previous step. However, for a branched route, the score is computed summing the scores of all the pathways connecting the route to the leaves. Each of these pathways is linear and is scored in the same way as described before.
-
(9)
The final step of the algorithm occurs after the search has been completed, and it can unfold in two different scenarios. In the first scenario, no solution has been found, and the system returns a failed status. In this case, no solution can be displayed to the user. In the second scenario, at least one solution has been found, and the algorithm proceeds with the finalization steps.
In this finalization step, an additional score value y is computed and returned to the user interface using the formula y = 1/(1 + log(x + 1)), where x is the tree-route score computed in the previous step. While arbitrary, the scoring function was chosen to return a number between 0 and 1, with an higher value connected to a greater confidence in the designed retrosynthesis.
Data availability
We provide sample data to run the method (https://github.com/rxn4chemistry/rxn-neb). The Pistachio and emolecules data are available in40 and45 respectively.
Code availability
The experiments presented in the paper can be reproduced using the RXN platform via UI (https://rxn.res.ibm.com) or REST API (Python client: https://github.com/rxn4chemistry/rxn4chemistry). Additionally, we provide and open source implementation of our algorithm that allows to run the method customizing: single-step forward/backward models, fingerprints model (https://github.com/rxn4chemistry/rxn-neb).
References
Corey, E. J. & Wipke, W. T. Computer-assisted design of complex organic syntheses. Science 166, 178–192 (1969).
Szymkuć, S. et al. Computer-assisted synthetic planning: the end of the beginning. Angew. Chem. Int. Ed. 55, 904–5937 (2016).
Liu, B. et al. Retrosynthetic reaction prediction using neural sequence-to-sequence models. ACS Cent. Sci. 3, 1103–1113 (2017).
Segler, M. H. S. & Waller, M. P. Neural-symbolic machine learning for retrosynthesis and reaction prediction. Chem. Eur. J. 23, 5966–5971 (2017).
Dai, H., Li, C., Coley, C. W., Dai, B. & Song, L. Retrosynthesis prediction with conditional graph logic network. arXiv https://doi.org/10.48550/arXiv.2001.01408 (2020).
Segler, M. H. S., Preuss, M. & Waller, M. P. Planning chemical syntheses with deep neural networks and symbolic AI. Nature 555, 604–610 (2018).
Chen, B., Shen, T., Jaakkola, T. S. & Barzilay, R. Learning to make generalizable and diverse predictions for retrosynthesis. arXiv https://doi.org/10.48550/arXiv.1910.09688 (2019).
Zheng, S., Rao, J., Zhang, Z., Xu, J. & Yang, Y. Predicting retrosynthetic reactions using self-corrected transformer neural networks. J. Chem. Inf. Model 60, 47–55 (2020).
Coley, C. W. A robotic platform for flow synthesis of organic compounds informed by AI planning. Science 365, eaax1566 (2019).
Schreck, J. S., Coley, C. W. & Bishop, K. J. M. Learning retrosynthetic planning through simulated experience. ACS Cent. Sci. 5, 970–981 (2019).
Baylon, J. L., Cilfone, N. A., Gulcher, J. R. & Chittenden, T. W. Enhancing retrosynthetic reaction prediction with deep learning using multiscale reaction classification. J. Chem. Inf. Model 59, 673–688 (2019).
Molga, K., Dittwald, P. & Grzybowski, B. A. Navigating around patented routes by preserving specific motifs along computer-planned retrosynthetic pathways. Chem 5, 460–473 (2019).
Lee, A. A. et al. Molecular transformer unifies reaction prediction and retrosynthesis across pharma chemical space. Chem. Commun. 55, 12152–12155 (2019).
Schwaller, P. et al. Predicting retrosynthetic pathways using transformer-based models and a hyper-graph exploration strategy. Chem. Sci. 11, 3316–3325 (2020).
Karpov, P., Godin, G. & Tetko, I. V. A transformer model for retrosynthesis. In International Conference on Artificial Neural Networks 817–830 (2019).
Ishida, S., Terayama, K., Kojima, R., Takasu, K. & Okuno, Y. Prediction and interpretable visualization of retrosynthetic reactions using graph convolutional networks. J. Chem. Inf. Model 59, 5026–5033 (2019).
Lin, K., Xu, Y., Pei, J. & Lai, L. Automatic retrosynthetic route planning using template-free models. Chem. Sci. 11, 3355–3364 (2020).
Shi, C., Xu, M., Guo, H., Zhang, M. & Tang, J. A graph to graphs framework for retrosynthesis prediction. In Proceedings of the 37th International Conference on Machine Learning 8818–8827 (2020).
Genheden, S. Aizynthfinder: a fast, robust and flexible open-source software for retrosynthetic planning. J. Cheminform. 12, 70 (2020).
Tetko, I. V., Karpov, P., Van Deursen, R. & Godin, G. State-of-the-art augmented NLP transformer models for direct and single-step retrosynthesis. Nat. Commun. 11, 5575 (2020).
Chen, B., Li, C., Dai, H. & Song, L. Retro*: learning retrosynthetic planning with neural guided A* search. In International Conference on Machine Learning, 1608–1616 (PMLR, 2020).
Mikulak-Klucznik, B. et al. Computational planning of the synthesis of complex natural products. Nature 588, 83–88 (2020).
Badowski, T., Gajewska, E. P., Molga, K. & Grzybowski, B. A. Synergy between expert and machine-learning approaches allows for improved retrosynthetic planning. Angew. Chem. Int. Ed. Engl. 59, 725–730 (2020).
Hasic, H. & Ishida, T. Single-step retrosynthesis prediction based on the identification of potential disconnection sites using molecular substructure fingerprints. J. Chem. Inf. Model 61, 641–652 (2021).
Amol Thakkar, A. et al. Artificial intelligence and automation in computer aided synthesis planning. React. Chem. Eng. 6, 27–51 (2021).
Mao, K. et al. Molecular graph enhanced transformer for retrosynthesis prediction. Neurocomputing 457, 193–202 (2021).
Wang, X. R. et al. Retroprime: a diverse, plausible and transformer-based method for single-step retrosynthesis predictions. Chem. Eng. J. 420, 129845 (2021).
Ishida, S., Terayama, K., Kojima, R., Takasu, K. & Okuno, Y. AI-driven synthetic route design incorporated with retrosynthesisknowledge. J. Chem. Inf. Model 62, 1357–1367 (2022).
Thakkar, A. et al. Unbiasing retrosynthesis language models with disconnection prompts. ACS Cent. Sci. 9, 1488 (2023).
Pasquini, M. & Stenta, M. LinChemIn: route arithmetic-operations on digital synthetic routes. J. Chem. Inf. Model. 64, 1765–1771 (2024).
Lin, M. H., Tu, Z. & Coley, C. W. Improving the performance of models for one-step retrosynthesis through re-ranking. J. Cheminform. 14, 15 (2022).
Warren, S. & Wyatt, P. Organic Synthesis: The Disconnection Approach 2nd ed. Wiley (2011).
Yu, Y. et al. GRASP: navigating retrosynthetic planning with goal-driven policy. Adv. Neural Inf. Process. Syst. 35, 10257–10268 (2022).
Zhong, Z. et al. Recent advances in deep learning for retrosynthesis. WIREs Comput. Mol. Sci. 14, e1694 (2023).
Jiang, Y. et al. Artificial intelligence for retrosynthesis prediction. Engineering 25, 32–50 (2023).
Schwaller, P. et al. Mapping the space of chemical reactions using attention-based neural networks. Nat. Mach. Intell. 3, 144–152 (2021).
Janet, J. P., Tomberg, A. & Bostrőm, J. Reusability report: Learning the language of synthetic methods used in medicinal chemistry. Nat. Mach. Intell. 3, 572–575 (2021).
Schwaller, P., Vaucher, A. C., Laino, T. & Reymond, J.-L. Prediction of chemical reaction yields using deep learning. Mach. Learn. Sci. Technol. 2, 015016 (2021).
Wang, X. et al. From theory to experiment: transformer-based generation enables rapid discovery of novel reactions. J. Cheminform. 14, 1–14 (2022).
Nextmove Software, Pistachio. https://www.nextmovesoftware.com/pistachio.html. Accessed 2021.
Schneider, N., Lowe, D. M., Sayle, R. A. & Landrum, G. A. Development of a novel fingerprint for chemical reactions and its application to large-scale reaction classification and similarity. J. Chem. Inf. Model. 55, 39–53 (2015).
Andraos, J. On using tree analysis to quantify the material, input energy, and cost throughput efficiencies of simple and complex synthesis plans and networks: towards a blueprint for quantitative total synthesis and green chemistry. Org. Process Res. Dev. 10, 212–240 (2006).
Weber, J. M., Lió, P. & Lapkin, A. A. Identification of strategic molecules for future circular supply chains using large reaction networks. React. Chem. Eng. 4, 1969–1981 (2019).
IBM RXN for chemistry. https://rxn.res.ibm.com. Accessed August 2023.
eMolecules Database. https://www.emolecules.com. Accessed August 29, 2019.
Schwaller, P. et al. Molecular transformer: a model for uncertainty-calibrated chemical reaction prediction. ACS Cent. Sci. 5, 1572–1583 (2019).
Acknowledgements
This publication was created as part of NCCR Catalysis (grant number 180544), a National Centre of Competence in Research funded by the Swiss National Science Foundation.
Author information
Authors and Affiliations
Contributions
T.L. laid the foundational groundwork for the project through the initial design and conceptualization of the idea. F.Z. initiated the conceptual development of the project, expanded and adapted the original idea, and was primarily responsible for implementing the string approach. C.B. provided expert analysis and critical evaluation of the retrosynthesis strategy, ensuring the robustness and reliability of the proposed chemical synthesis. M.M. gave insights technical execution of the project, directly contributing to the implementation and integration of the project with the RXN platform. M.M. oversaw the process of open-sourcing of the project code and the assembling the scripts for file processing for easy accessibility and applicability of the work. J.B. was responsible for processing the data from the Pistachio and USPTO databases. T.L. also evaluated the retrosynthesis strategy, contributing to the strategic direction and methodological rigor of the research. All authors contributed significantly to the manuscript, collaborating on the drafting and critical revision of the text to ensure the clarity, accuracy, and coherence of the final publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zipoli, F., Baldassari, C., Manica, M. et al. Growing strings in a chemical reaction space for searching retrosynthesis pathways. npj Comput Mater 10, 101 (2024). https://doi.org/10.1038/s41524-024-01290-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41524-024-01290-x