Abstract
Energy imbalance has an important role in breast cancer prognosis. Hyperactive mechanistic Target of Rapamycin (mTOR) pathway is associated with breast tumor growth, but the extent to which body fatness is associated with mTOR pathway activities in breast cancer is unclear. We performed immunostaining for mTOR, phosphorylated (p)-mTOR, p-AKT, and p-p70S6K in tumor tissue from 590 women (464 African Americans/Blacks and 126 Whites) with newly diagnosed invasive breast cancer in the Women’s Circle of Health Study. Anthropometric measures were taken by study staff, and body composition was measured by bioelectrical impedance analysis. Linear regressions were used to estimate percent differences in protein expression between categories of body mass index (BMI), waist circumference, waist/hip ratio, fat mass, fat mass index, and percent body fat. We observed that BMI ≥ 35.0 vs. <25 kg/m2 was associated with 108.3% (95% CI = 16.9%–270.9%) and 101.8% (95% CI = 17.0%–248.8%) higher expression in p-mTOR and normalized p-mTOR, i.e., p-mTOR/mTOR, respectively. Quartiles 4 vs. 1 of waist/hip ratio was associated with 41.8% (95% CI = 5.81%–89.9%) higher mTOR expression. Similar associations were observed for the body fat measurements, particularly in patients with estrogen receptor-negative (ER−) tumors, but not in those with ER+ tumors, although the differences in associations were not significant. This tumor-based study found positive associations between body fatness and mTOR pathway activation, evident by a p-mTOR expression, in breast cancer. Our findings suggest that mTOR inhibition can be a treatment strategy to prevent the recurrence of these tumors in obese individuals.
Similar content being viewed by others
Introduction
Energy imbalance has an important role in breast cancer prognosis: compared with individuals with normal weight (18.5–24.99 kg/m2) at diagnosis, those with obesity (body mass index [BMI] ≥ 30 kg/m2) are associated with a 30% increased risk of all-cause or breast cancer-specific mortality, regardless of menopausal status or tumor estrogen receptor (ER) status1,2,3. The aromatization process in adipose tissue produces estrogens that promote ER+ cancer; however, the hormonal pathway cannot fully account for the association because the association is also observed in patients who receive tamoxifen or aromatase inhibitors4,5 and in patients with ER− tumors1. In addition, although the evidence is less compared to BMI, central adiposity is associated with poor prognosis in patients with breast cancer6, and the mechanism between central adiposity and outcomes may be different from that of BMI and outcome. Although some clinical trials examining behavioral changes (e.g., weight reduction through a decreased fat intake and increased physical activity) reveal promising findings7,8, it is crucial to identify mechanisms through which overall and central adiposity exert their effects. Lifestyle interventions may not be applicable or effective for all women with breast cancer; targeting the underlying biological mechanisms may open new opportunities to improve the prognosis for a greater number of patients.
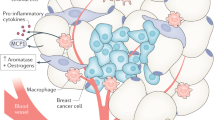
An obesity-related signaling pathway is a mechanistic Target of Rapamycin (mTOR) pathway (Fig. 1). The mTOR pathway is activated by energy influx, amino acids, and insulin-like growth factors (IGFs)9, and activation of the pathway promotes several cancer hallmarks, such as cell proliferation and angiogenesis10,11. and is associated with an increased risk of breast cancer recurrence12. The main signaling mechanism of the pathway is protein phosphorylation, a post-translational process11. Phosphorylated mTOR and its upstream and downstream proteins, such as p70S6 kinase (p70S6K), are highly expressed in breast tumors13,14,15,16. However, data are very limited on the association between obesity phenotypes and mTOR pathway activation in breast tumors. This knowledge would advance our understanding of the mechanism of how energy imbalance affects breast cancer prognosis and shed light on the potential for promoting energy balance and mTOR inhibition as strategies to improve clinical outcomes17,18.
*Protein expression was assessed using IHC in this study. 4E-BP1 4E-binding protein-1, AKT Protein kinase B, eIF-4E eukaryotic initiation factor-4E, IRS insulin receptor substrate, mTORC1, and mTORC2 mTOR Complex 1 and 2, PI3K phosphoinositide 3-kinases, PRAS40 proline-rich Akt substrate 40 kDa, PTEN phosphatase and tensin homolog, S6K1 S6 kinase-1 (also known as p70S6).
Here, we investigated the mTOR pathway activities in breast tumors in association with body size, i.e., BMI, waist circumference (WC), and waist/hip ratio (WHR), as well as body composition, i.e., fat mass, fat mass index, and percent body fat. We hypothesized that mTOR pathway activities would be higher in women with larger body size and more fat mass, compared to those with smaller body size and less fat mass. Because the influence of obesity may differ between the ER+ and ER− subtypes of breast cancer19,20, we also examined the mTOR-body fatness associations stratified by ER status.
Results
Protein expression and patient characteristics
The distributions of H-scores for IHC markers are listed in Table 1. mTOR and p-mTOR expression levels were modestly correlated (r = 0.35; Supplementary Table 1). The expression levels of phosphoproteins, i.e., p-mTOR, p-AKT, and p-p70S6K were also modestly correlated with each other (r = 0.27–0.54). Table 2 shows the univariate analysis of the protein expression levels of mTOR, p-mTOR, normalized p-mTOR, and total phosphoproteins according to demographic and clinicopathological characteristics. mTOR expression was higher in tumors from Black than White women (median H-score = 145 vs. 96, P < 0.001). Patients with a tumor of lower grade and smaller size and an earlier stage of breast cancer had higher expression levels of p-mTOR and total phosphoproteins than those with a tumor of higher grade and larger size and a later stage of the disease. The expression levels of mTOR, p-mTOR, and total phosphoproteins were higher in ER+ and PR+ than ER− and PR− tumors; the expression levels were lowest in triple-negative tumors among the molecular subtypes. In multivariable models, the Black race compared to the White race was significantly associated with higher expression of mTOR and p-mTOR, but not normalized p-mTOR or total phosphoproteins, in breast tumors after adjusting for age, menopausal status, history of diabetes, grade, stage, molecular subtype, and BMI category (Supplementary Table 2).
Body fatness measurements
Among the body fatness measurements, BMI was highly correlated with WC (r = 0.88), fat mass (r = 0.93), fat mass index (r = 0.97), and percent body fat (r = 0.80) (Supplementary Table 3). WHR was modestly correlated with WC (r = 0.57), and BMI (r = 0.34). The correlations were high (r = 0.91–0.98) among the fat mass, fat mass index, and percent body fat.
Body fatness and protein expression
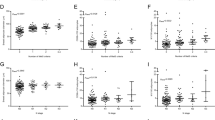
BMI was positively associated with greater expression of p-mTOR and normalized p-mTOR (P-trend = 0.011 and 0.046, respectively; Table 3). Tumors from women with BMI ≥ 35 vs. those with BMI < 25 kg/m2 had 108.3% (95% CI = 16.9%–270.9%, P = 0.012) higher level of p-mTOR expression and 101.8% (95% CI = 17.0%–248.8%, P = 0.012) higher level of normalized p-mTOR expression. Quartiles 4 vs. 1 of WHR was associated with 41.8% (95% CI = 5.81%–89.9%, P = 0.019, P-trend = 0.023) higher level of mTOR expression. For body composition measurements (Table 4), higher vs. lower fat mass, fat mass index, and percent body fat were associated with greater p-mTOR and normalize p-mTOR expression (all P-trend <0.05). Quartiles 4 vs. 1 of fat mass, fat mass index, and percent body fat were associated with 149.8%, 124.1%, and 123.9% higher levels of p-mTOR expression, respectively (all P < 0.05). The estimates were similar for normalized p-mTOR expression. We observed a lower mTOR expression among women with overweight (vs. normal weight) and those in the second quartile of percent body fat (vs. the first quartile). However, these associations were not observed for the phosphoproteins. There was no association of total phosphoprotein, p-AKT, or p-p70S6K (Supplementary Table 4) with the body fatness measurements.
We examined the associations by ER status for BMI and percent body fat in relation to the expression of p-mTOR and normalized p-mTOR (Table 5). BMI ≥ 35 vs. <25 kg/m2 was associated with a higher level of normalized p-mTOR expression in ER− tumor (191.2%, 95% CI = 10.5%–667.9%, P = 0.031), but not in ER+ tumors. Similarly, Q4 vs. Q1 of percent body fat was associated with higher levels of p-mTOR and normalized p-mTOR expression in ER− tumors (207.6% and 283.3%, respectively, both P < 0.05), but not in ER+ tumors. These differences in associations between ER− and ER+ tumors were not statistically significant (P-heterogeneity >0.05).
Discussion
In this study population comprised predominantly of Black women, we used a panel of IHC protein and phosphoproteins to indicate mTOR pathway activities. Among the assayed proteins, p-mTOR, both the original expression level and the normalized level, was associated with BMI and body fat mass. There was a pattern that the associations were more prominent in ER− than ER+ tumors, although the differences in associations between the subtypes were not significant.
Our study is among the few reporting the association between body fatness and mTOR pathway protein expression in breast tumors. Prior research reported obesity-related gene expression signatures in breast tumors21,22. Creighton et al.22 reported that gene expression of IGF-related proteins, which promote the mTOR pathway, was higher in obese verse normal weight breast cancer patients (N = 103). Fuentes-Mattei et al.21 observed significant upregulation of AKT/mTOR genes in obese vs. normal-weight women with ER+ tumors (N = 137). However, these findings were not replicated in a study of 519 postmenopausal women23. As obesity may affect both gene and protein expression in tumors, we focus on the latter because the mTOR pathway signaling is primarily through post-translational protein phosphorylation24,25. In our data, p-mTOR, but not p-AKT or p-p70S6K, in breast tumors was associated with body fatness, despite that one would expect these markers to yield similar results and they were modestly correlated. The discrepancies can be explained by the promotors of these proteins. mTOR is promoted by not only growth factors but also directly by nutrients and energy. AKT is activated by IGF1; however, IGF1 is not linearly associated with body fatness as the blood levels of IGF1 decreases when BMI ≥ 27 kg/m2 26. Also, AKT signaling relies on the status of other proteins such as PI3K and PTEN. p70S6K can be signaled independently from mTOR, as it is downstream of phosphoinositide-dependent kinase-1 (PDK1) in the IGF signaling pathway27 and the ER signaling pathway28. Additional markers are needed to more comprehensively assess the singling pathways in breast cancer in relation to body fatness.
In the U.S., Black women with breast cancer have consistently lower survival than White women, despite a lower incidence of breast cancer until recent years29,30. In addition to societal and socioeconomic factors, such as access to screening and receipt of optimal treatment, tumor characteristics, e.g., grade, stage, and breast cancer subtype31, and molecular features32,33,34,35,36 likely contribute to differences in breast cancer mortality rates. We observed that the mTOR and p-mTOR expression were higher in Black women than White women after adjusting for tumor characteristics and BMI. The findings suggest that biological factors, such as race-related differences in insulin resistance37, may have a role in the racial disparity in the mTOR pathway activities and potentially survival. A limitation of our observation is that the sample size of White women was relatively small to Black women; the finding warrants replication.
In this study, mTOR pathway activities were higher in ER+ than ER− tumors. The finding is consistent with the biological evidence that there is a crosstalk between the ER and mTOR signaling pathways38,39. Nevertheless, it has been hypothesized that for ER− tumors, the influence of obesity may be more related to insulin signaling than estrogens40,41. Our observation on the associations of BMI and percent body fat with p-mTOR expression in ER− tumors supports the hypothesis. The sample size of women with ER− tumors was relatively small compared to ER+ tumors and the estimates had wide 95% confidence intervals. Thus, our findings in ER− tumors should be interpreted with caution and warrant confirmation. Clinically, it is important to know whether mTOR activation is related to the recurrence of these tumors in obese individuals and whether mTOR inhibition is helpful to prevent recurrence. mTOR inhibition can be achieved with mTOR inhibitors or metformin, an insulin sensitizer that is widely used in the treatment of type II diabetes42,43. Metformin reduces breast tumor growth in diet-induced obese mice44. In humans, metformin as adjuvant therapy with weight loss was found to be a safe strategy that modestly lowered circulating estrogen and insulin levels45. A large clinical trial (Canadian Cancer Trials Group’s MA.32) is ongoing to reveal the effect of metformin on breast cancer prognosis and survival46.
Other strengths of our study include rigorous and quantitative measurements of protein expression in breast tumors and body fatness. Immunostaining on FFPE tissue samples remains a cost-effective option for the assessment of protein expression in large epidemiologic studies, in which fresh-frozen tissue samples are unavailable. The stained tissue was manually annotated so that influences from other tissue components (e.g. stroma) on scoring were minimized. In the annotated images, using automated imaging analysis gives objective scores for each localization. Also, body size and body composition were measured by trained staff with a standardized protocol.
Limitations of our study should be noted. The study population was breast cancer cases from a case-control study, and body size and body composition might have been influenced by breast cancer development. In addition, body fatness might have been affected by breast cancer treatment because body fatness was measured within nine months after the time of breast cancer diagnosis and surgery. Research has shown a mean weight gain of 2 to 3 kg at 6 months to 1 year of chemotherapy in breast cancer patients47,48, while the weight gain is less for patients receiving surgery and radiation only49. Although we performed planned analyses with a priori hypotheses, the investigation of different measurements of body fatness resulted in multiple comparisons, potentially leading to false-positive results. Another limitation is that our 4-marker protein expression panel may have missed important signaling of the mTOR pathway activation. Future studies should measure additional proteins and mutations in the pathway for a more comprehensive assessment.
In conclusion, our study showed positive associations between body fatness and mTOR pathway activation, evident by a p-mTOR expression, in breast cancer. Our findings suggest that mTOR inhibition should be studied as a treatment strategy or as a method to prevent the recurrence of these tumors in obese individuals.
Methods
Study population
Study participants were breast cancer cases based on the Women’s Circle of Health Study (WCHS), a multi-site case-control study in New York City and New Jersey designed to investigate genetic and lifestyle risk factors for aggressive breast cancer. Details on study recruitment have been described elsewhere50,51. The protocol was approved by all relevant Institutional Review Boards. In brief, cases were self-identified Black and White women, 20–75 years of age, with no previous history of cancer other than non-melanoma skin cancer, and diagnosed within 9 months of ascertainment with primary, histologically confirmed, invasive breast cancer or ductal carcinoma in situ (DCIS). In-home interviews were conducted to obtain questionnaire data on known and suspected risk factors for breast cancer and anthropometric measurements. Of participants eligible for inclusion in this analysis, more than 95% signed a release form for their tumor tissue, as part of the informed consent. Formalin-fixed paraffin-embedded (FFPE) tissue specimens were used for tissue microarray (TMA) construction that was guided by a specialized breast pathologist (T.K.). TMAs included two to three tumor cores ranging in size from 0.6 to 1.2 mm. A total of 770 cases had tissue samples on a TMA for laboratory assays. After immunostaining, DCIS cases (n = 68) and cases that did not have any tumor tissue cores with sufficient cells (<25 cells) for scoring were excluded. The final data set consisted of 590 cases, including 464 black women (299 ER+ cases and 165 ER− cases) and 126 white women (97 ER+ cases and 29 ER− cases), who had data on all four assayed mTOR pathway proteins. Clinical and tumor characteristics, including the expression status of hormone receptors (HR, i.e., ER and progesterone receptor [PR]) and human epidermal growth factor receptor 2 (HER2), were obtained from the pathology reports.
Ethics
All participants provided written informed consent to take part in the study. The protocol was approved by the Institutional Review Boards of all participating institutions, including Roswell Park Comprehensive Cancer Center and Rutgers Cancer Institute of New Jersey.
Immunohistochemistry and image analysis
TMAs were sectioned at 5 µm and stained by immunohistochemistry (IHC) methods for mTOR (clone 7C10), phosphorylated mTOR (p-mTOR, Ser2448), phosphorylated AKT (p-AKT, Ser4731), and phosphorylated P70S6K (p-p70S6K, T389). The antibodies and IHC conditions for each stain are listed in Supplementary Table 5. Stained slides were digitally imaged at ×20 magnification using the Aperio ScanScope XT (Leica Biosystems), and the images were manually annotated to identify tumor for analysis. Automated image analysis on the annotated regions was performed using Aperio algorithms with minor adjustments for cell shape and intensity thresholds. The algorithms were validated by our study pathologist (W.B.) using positive and negative controls and tissue samples with different levels of intensity. Tumor cell cytoplasm was scored for mTOR and p-mTOR; cytoplasm and nuclei were scored for p-AKT and p-p70S6K. The percentage of cells stained was recorded in each intensity category: 0 (no staining), 1+ (only partial or weak staining), 2+ (moderate staining), and 3+ (strong staining). The core-level data were collapsed into patient-level data using a cellularity-weighted approach52. Core weight was defined as the number of tumor cells in a given core divided by the total number of tumor cells across all cores for that patient. The weighted average at the patient level was calculated by summing the product of percent positivity in each of the intensity category and core weight across all cores per patient. With the weighted average of percent positivity values, a histological score (H-score) at the patient level was calculated by the formula: [1 × (% cells 1+) + 2 × (% cells 2+) + 3 × (% cells 3+)] × 10053. Figure 2 show representative images of IHC staining.
Anthropometric and body composition measurements
Anthropometric measurements were taken by trained staff during the in-person interviews using a standardized protocol described elsewhere54. Participants were asked to wear light clothing and to remove their shoes and any heavy jewelry. Waist and hip circumferences were measured by placing the measuring tape around the waist covering the umbilicus for waist and at the maximum extension of the buttocks in a horizontal plane for the hip. The waist and hip measurements were taken twice to the nearest 0.1 cm. If the difference between the first and second measurement was greater than 2 cm, a third measurement was taken. The two (or three) measurements were averaged for analyses. Standing height was measured once to the nearest 0.1 cm. Body composition was measured by bioelectrical impedance analysis using a Tanita® TBF-300A scale. Weight was measured once using the Tanita scale. BMI was classified as <25.0 (normal weight), 25.0–29.9 (overweight), 30.0–34.9 (class I obesity), and ≥35.0 kg/m2 (class II/III obesity)55. The fat mass index was calculated as a fat mass in kg divided by the square of height in meters.
Statistical analysis
We examined the associations between each assayed protein and the body fatness measurements. In addition, because p-mTOR was stained for the specific phosphorylated site of mTOR and the levels of p-mTOR expression may depend on the expression of mTOR, we normalized the p-mTOR expression as the H-score of p-mTOR divided by the H-score of mTOR multiplied by 100. In addition, an H-score of total phosphoprotein was derived as the summation of H-scores from p-mTOR, p-AKT, and p-p70S6K to better represent the mTOR pathway activities than using single markers. Correlations of H-scores between individual proteins and between body fatness measurements were examined using the Pearson correlation coefficient. The protein expression levels were examined by demographic and clinicopathological characteristics using the analysis of variance (ANOVA). Linear regression was performed to assess the associations between the body fatness measurements, i.e., the independent variables and protein expression levels, i.e., the dependent variables. Because the distributions of H-scores were right-skewed, the data were log-transformed to improve the normality for the regression analysis. The regression coefficient (β) was converted to percentage using the formula: (exp (β) − 1) × 100% and interpreted as estimated percentage difference of protein expression for each category compared with the reference category. The BMI categories, as well as the quartiles of WC, WHR, fat mass, fat mass index, and percent body fat, were entered as categorical variables in regression models. Quartile 1 or BMI < 25 kg/m2 was chosen as the reference group. Other covariates were race (Black or White), menopausal status (premenopausal or postmenopausal), history of diabetes (ever or never), and breast cancer molecular subtype (HR+/HER2−, HR+/HER2+, HR−/HER2+, or HR−/HER2−)56. The effects of breast cancer stage and tumor grade were largely explained by molecular subtype and thus not included in the final model. Because the body size and composition measurements were highly correlated, only one measurement was included in a model to avoid multicollinearity. P-values for trend were obtained by treating the categories of body fatness measurement as continuous variables in the regression models. As planned, a stratification analysis was performed according to the ER status of breast cancer. To reduce the number of comparisons, only p-mTOR and normalized p-mTOR were examined for BMI and percent body fat in the stratification analysis, as the two proteins showed consistent results with BMI and the body fat variables in the main analysis. The difference in associations between ER+ and ER− tumors were examined using the contrast test method; p-values for heterogeneity were reported57. All tests of statistical significance were two-sided; a P-value < 0.05 was considered statistically significant. All analyses were planned and the results were not adjusted for multiplicity. Statistical analyses were performed using SAS, version 9.4 (SAS Institute Inc).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data generated and analyzed during this study are described in the following metadata record: https://doi.org/10.6084/m9.figshare.1275258258. The data supporting the findings of this study are not publicly available in order to protect patient privacy. The data will be made available to authorized researchers who have submitted an IRB application. Please apply to the Women’s Circle of Health Study (WCHS) committee, email address: tingyuan.cheng@ufl.edu, for access to this data set.
Code availability
Aperio (Leica Biosystems) was utilized in the automatic image analysis. The nuclear and cytoplasm algorithms can be available upon request.
References
Niraula, S., Ocana, A., Ennis, M. & Goodwin, P. J. Body size and breast cancer prognosis in relation to hormone receptor and menopausal status: a meta-analysis. Breast Cancer Res. Treat. 134, 769–781 (2012).
Protani, M., Coory, M. & Martin, J. H. Effect of obesity on survival of women with breast cancer: systematic review and meta-analysis. Breast Cancer Res. Treat. 123, 627–635 (2010).
Chan, D. S. et al. Body mass index and survival in women with breast cancer-systematic literature review and meta-analysis of 82 follow-up studies. Ann. Oncol. 25, 1901–1914 (2014).
Sestak, I. et al. Effect of body mass index on recurrences in tamoxifen and anastrozole treated women: an exploratory analysis from the ATAC trial. J. Clin. Oncol. 28, 3411–3415 (2010).
Ewertz, M. et al. Obesity and risk of recurrence or death after adjuvant endocrine therapy with letrozole or tamoxifen in the breast international group 1-98 trial. J. Clin. Oncol. 30, 3967–3975 (2012).
Bandera, E. V., Maskarinec, G., Romieu, I. & John, E. M. Racial/ethnic disparities in the impact of obesity on breast cancer risk and survival: A global perspective. Adv. Nutr. 6, 803–819 (2015).
Chlebowski, R. T. & Reeves, M. M. Weight loss randomized intervention trials in female cancer survivors. J. Clin. Oncol. 34, 4238–4248 (2016).
Irwin, M., Fabian, C. & McTiernan, A. in Improving Outcomes for Breast Cancer Survivors (ed. Ganz, P. A.) 193–212 (Breast Cancer Research Foundation, Springer, 2015).
Renehan, A. G. et al. Insulin-like growth factor (IGF)-I, IGF binding protein-3, and cancer risk: systematic review and meta-regression analysis. Lancet 363, 1346–1353 (2004).
Zoncu, R., Efeyan, A. & Sabatini, D. M. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 (2011).
Saxton, R. A. & Sabatini, D. M. mTOR signaling in growth, metabolism, and disease. Cell 169, 361–371 (2017).
Zhang, Y. et al. A pan-cancer proteogenomic atlas of PI3K/AKT/mTOR pathway alterations. Cancer Cell 31, 820–832 e823 (2017).
Beca, F. et al. p-mTOR expression is associated with better prognosis in luminal breast carcinoma. J. Clin. Pathol. 67, 961–967 (2014).
Beelen, K. et al. Phosphorylated p-70S6K predicts tamoxifen resistance in postmenopausal breast cancer patients randomized between adjuvant tamoxifen versus no systemic treatment. Breast Cancer Res. 16, R6 (2014).
Chiang, G. G. & Abraham, R. T. Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J. Biol. Chem. 280, 25485–25490 (2005).
Sonnenblick, A., Venet, D., Brohee, S., Ponde, N. & Sotiriou, C. pAKT pathway activation is associated with PIK3CA mutations and good prognosis in luminal breast cancer in contrast to p-mTOR pathway activation. NPJ Breast Cancer 5, 7 (2019).
De Angel, R. E. et al. The enhancing effects of obesity on mammary tumor growth and Akt/mTOR pathway activation persist after weight loss and are reversed by RAD001. Mol. Carcinog. 52, 446–458 (2013).
Goodwin, P. J. & Stambolic, V. Obesity and insulin resistance in breast cancer–chemoprevention strategies with a focus on metformin. Breast 20, S31–S35 (2011).
Althuis, M. D. et al. Etiology of hormone receptor-defined breast cancer: a systematic review of the literature. Cancer Epidemiol. Biomark. Prev. 13, 1558–1568 (2004).
Picon-Ruiz, M., Morata-Tarifa, C., Valle-Goffin, J. J., Friedman, E. R. & Slingerland, J. M. Obesity and adverse breast cancer risk and outcome: mechanistic insights and strategies for intervention. CA Cancer J. Clin. 67, 378–397 (2017).
Fuentes-Mattei, E. et al. Effects of obesity on transcriptomic changes and cancer hallmarks in estrogen receptor-positive breast cancer. J. Natl Cancer Inst. 106, dju158 (2014).
Creighton, C. J. et al. A gene transcription signature of obesity in breast cancer. Breast Cancer Res. Treat. 132, 993–1000 (2012).
Heng, Y. J. et al. Molecular mechanisms linking high body mass index to breast cancer etiology in post-menopausal breast tumor and tumor-adjacent tissues. Breast Cancer Res. Treat. 173, 667–677 (2019).
Laplante, M. & Sabatini, D. M. mTOR signaling in growth control and disease. Cell 149, 274–293 (2012).
Day, E. K., Sosale, N. G. & Lazzara, M. J. Cell signaling regulation by protein phosphorylation: a multivariate, heterogeneous, and context-dependent process. Curr. Opin. Biotechnol. 40, 185–192 (2016).
Endogenous Hormones Breast Cancer Collaborative Group, Key, T. J., Appleby, P. N., Reeves, G. K. & Roddam, A. W. Insulin-like growth factor 1 (IGF1), IGF binding protein 3 (IGFBP3), and breast cancer risk: pooled individual data analysis of 17 prospective studies. Lancet Oncol. 11, 530–542 (2010).
Harada, H., Andersen, J. S., Mann, M., Terada, N. & Korsmeyer, S. J. p70S6 kinase signals cell survival as well as growth, inactivating the pro-apoptotic molecule BAD. Proc. Natl Acad. Sci. USA 98, 9666–9670 (2001).
Berman, A. Y. et al. ERRalpha regulates the growth of triple-negative breast cancer cells via S6K1-dependent mechanism. Signal Transduct. Target Ther. 2, e17035 (2017).
Richardson, L. C., Henley, S. J., Miller, J. W., Massetti, G. & Thomas, C. C. Patterns and trends in age-specific black-white differences in breast cancer incidence and mortality - United States, 1999-2014. MMWR Morb. Mortal. Wkly Rep. 65, 1093–1098 (2016).
DeSantis, C. E., Ma, J., Goding Sauer, A., Newman, L. A. & Jemal, A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J. Clin. 67, 439–448 (2017).
Iqbal, J., Ginsburg, O., Rochon, P. A., Sun, P. & Narod, S. A. Differences in breast cancer stage at diagnosis and cancer-specific survival by race and ethnicity in the United States. JAMA 313, 165–173 (2015).
Keenan, T. et al. Comparison of the genomic landscape between primary breast cancer in African American versus White Women and the association of racial differences with tumor recurrence. J. Clin. Oncol. 33, 3621–3627 (2015).
Ambrosone, C. B. et al. Genome-wide methylation patterns provide insight into differences in breast tumor biology between American women of African and European ancestry. Oncotarget 5, 237–248 (2014).
Huo, D. et al. Comparison of breast cancer molecular features and survival by african and european ancestry in The Cancer Genome Atlas. JAMA Oncol. 3, 1654–1662 (2017).
Troester, M. A. et al. Racial differences in PAM50 subtypes in the Carolina Breast Cancer Study. J. Natl Cancer Inst. 110, 176–182 (2018).
Warner, E. T. et al. Racial and ethnic differences in breast cancer survival: mediating effect of tumor characteristics and sociodemographic and treatment factors. J. Clin. Oncol. 33, 2254–2261 (2015).
Hasson, R. E. & Goran, M. I. in Impact of Energy Balance on Cancer Disparities (eds. Bowen, D. J., Denis, G. V. & Berger, N. A.) 165–199 (Springer International Publishing, Switzerland, 2014).
Shrivastav, A. et al. The mechanistic target for rapamycin pathway is related to the phosphorylation score for estrogen receptor-alpha in human breast tumors in vivo. Breast Cancer Res. 16, R49 (2014).
Ciruelos Gil, E. M. Targeting the PI3K/AKT/mTOR pathway in estrogen receptor-positive breast cancer. Cancer Treat. Rev. 40, 862–871 (2014).
Davis, A. A. & Kaklamani, V. G. Metabolic syndrome and triple-negative breast cancer: a new paradigm. Int J. Breast Cancer 2012, 809291 (2012).
Dietze, E. C., Sistrunk, C., Miranda-Carboni, G., O’Regan, R. & Seewaldt, V. L. Triple-negative breast cancer in African-American women: disparities versus biology. Nat. Rev. Cancer 15, 248–254 (2015).
DeCensi, A. et al. Effect of metformin on breast ductal carcinoma in situ proliferation in a randomized presurgical trial. Cancer Prev Res. 8, 888–894 (2015).
Dowling, R. J. et al. Changes in insulin receptor signaling underlie neoadjuvant metformin administration in breast cancer: a prospective window of opportunity neoadjuvant study. Breast Cancer Res. 17, 32 (2015).
Kolb, R. et al. Obesity-associated NLRC4 inflammasome activation drives breast cancer progression. Nat. Commun. 7, 13007 (2016).
Patterson, R. E. et al. The effects of metformin and weight loss on biomarkers associated with breast cancer outcomes. J. Natl Cancer Inst. 110, 1239–1247 (2018).
Goodwin, P. J. et al. Effect of metformin vs placebo on weight and metabolic factors in NCIC CTG MA.32. J. Natl Cancer Inst. 107, djv006 (2015).
Demark-Wahnefried, W. et al. Changes in weight, body composition, and factors influencing energy balance among premenopausal breast cancer patients receiving adjuvant chemotherapy. J. Clin. Oncol. 19, 2381–2389 (2001).
Harvie, M. N., Campbell, I. T., Baildam, A. & Howell, A. Energy balance in early breast cancer patients receiving adjuvant chemotherapy. Breast Cancer Res. Treat. 83, 201–210 (2004).
Vance, V., Mourtzakis, M., McCargar, L. & Hanning, R. Weight gain in breast cancer survivors: prevalence, pattern and health consequences. Obes. Rev. 12, 282–294 (2011).
Ambrosone, C. B. et al. Conducting molecular epidemiological research in the age of HIPAA: a multi-institutional case-control study of breast cancer in African-American and European-American Women. J. Oncol. https://doi.org/10.1155/2009/871250 (2009).
Bandera, E. V. et al. Rethinking sources of representative controls for the conduct of case-control studies in minority populations. BMC Med. Res. Methodol. 13, 71 (2013).
Allott, E. H. et al. Performance of three-biomarker immunohistochemistry for intrinsic breast cancer subtyping in the AMBER Consortium. Cancer Epidemiol. Biomark. Prev. 25, 470–478 (2016).
Pirker, R. et al. EGFR expression as a predictor of survival for first-line chemotherapy plus cetuximab in patients with advanced non-small-cell lung cancer: analysis of data from the phase 3 FLEX study. Lancet Oncol. 13, 33–42 (2012).
Bandera, E. V. et al. Body fatness and breast cancer risk in women of African ancestry. BMC Cancer 13, 475 (2013).
Expert panel on the identification, evaluation, and treatment of overweight in adults. Clinical guidelines on the identification, evaluation, and treatment of overweight and obesity in adults: executive summary. Am. J. Clin. Nutr. 68, 899–917 (1998).
Allott, E. H. et al. Frequency of breast cancer subtypes among African American women in the AMBER consortium. Breast Cancer Res. 20, 12 (2018).
Wang, M. et al. Statistical methods for studying disease subtype heterogeneity. Stat. Med. 35, 782–800 (2016).
Cheng, T. D. et al. Metadata supporting the published article: body fatness and mTOR pathway activation of breast cancer in the Women’s Circle of Health Study. figshare https://doi.org/10.6084/m9.figshare.12752582 (2020).
Acknowledgements
This work was supported by grants from the US National Institutes of Health (P01 CA151135, R01 CA100598, R01 CA185623, P30 CA016056, P30 CA072720, R25 CA203650, and K07 CA201334), US Army Medical Research and Material Command (DAMD-17-01-1-0334), the Breast Cancer Research Foundation (C.B.A.; C.-C.H.), and a gift from the Philip L. Hubbell family.
Author information
Authors and Affiliations
Contributions
Study conception and design: T.Y.C. and C.B.A. Data acquisition: A.R.O, P.V.S., L.Z.P., W.B., R.P.O., W.D., C.C.H., E.V.B., T.K., and C.B.A. Writing—initial draft: T.Y.C. Data analysis: T.Y.C., R.P.O., W.Z., and S.L. Data interpretation: T.Y.C., A.R.O., S.Y., W.Z., S.D., and C.B.A., writing—final review and approval: T.Y.C., A.R.O., S.Y., W.Z., and C.B.A.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cheng, TY.D., Omilian, A.R., Yao, S. et al. Body fatness and mTOR pathway activation of breast cancer in the Women’s Circle of Health Study. npj Breast Cancer 6, 45 (2020). https://doi.org/10.1038/s41523-020-00187-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41523-020-00187-4
This article is cited by
-
Exercise interventions on body composition and quality of life of overweight/obese breast cancer survivors: a meta-analysis
BMC Women's Health (2023)
-
mTOR pathway candidate genes and energy intake interaction on breast cancer risk in Black women from the Women’s Circle of Health Study
European Journal of Nutrition (2023)
-
Results and lessons from dual extraction of DNA and RNA from formalin-fixed paraffin-embedded breast tumor tissues for a large Cancer epidemiologic study
BMC Genomics (2022)
-
mTOR pathway gene expression in association with race and clinicopathological characteristics in Black and White breast cancer patients
Discover Oncology (2022)
-
Body fatness and breast cancer risk in relation to phosphorylated mTOR expression in a sample of predominately Black women
Breast Cancer Research (2021)