Abstract
Cdk8 in Drosophila is the orthologue of vertebrate CDK8 and CDK19. These proteins have been shown to modulate transcriptional control by RNA polymerase II. We found that neuronal loss of Cdk8 severely reduces fly lifespan and causes bang sensitivity. Remarkably, these defects can be rescued by expression of human CDK19, found in the cytoplasm of neurons, suggesting a non-nuclear function of CDK19/Cdk8. Here we show that Cdk8 plays a critical role in the cytoplasm, with its loss causing elongated mitochondria in both muscles and neurons. We find that endogenous GFP-tagged Cdk8 can be found in both the cytoplasm and nucleus. We show that Cdk8 promotes the phosphorylation of Drp1 at S616, a protein required for mitochondrial fission. Interestingly, Pink1, a mitochondrial kinase implicated in Parkinson’s disease, also phosphorylates Drp1 at the same residue. Indeed, overexpression of Cdk8 significantly suppresses the phenotypes observed in flies with low levels of Pink1, including elevated levels of ROS, mitochondrial dysmorphology, and behavioral defects. In summary, we propose that Pink1 and Cdk8 perform similar functions to promote Drp1-mediated fission.
Similar content being viewed by others
Introduction
Cyclin-dependent kinase 19 (CDK19 [MIM: 614720]) and its paralog, CDK8 (MIM: 603184), are members of the transcriptional family of CDKs. Unlike other CDKs, these CDKs are not directly involved in cell-cycle regulatory processes1. Both CDK19 and CDK8 interact with Cyclin C to form a conserved kinase module that regulates RNA polymerase II-mediated transcription in the Mediator protein complex2. Enhanced CDK8 activity is implicated in tumorigenesis, including in colorectal cancer, breast cancer, and melanoma3,4,5,6.
We and others recently showed that mutations in CDK8/CDK19 are associated with severe neurodevelopmental disorders in humans7,8,9,10. Most patients with CDK19 pathogenic variants exhibit epilepsy, including infantile spasms, hypotonia, ataxia, developmental delay/severe intellectual disability, central nervous system (CNS) changes on MRI (brain atrophy, delayed myelination), and craniofacial dysmorphisms. Flies that lack cdk8 in neurons, the orthologue of CDK8/CDK19, exhibit similar phenotypes such as reduced lifespan and susceptibility to seizures9. Most de novo variants identified in CDK8 and CDK19 map to the ATP-binding domain associated with the kinase activity7,8,9,10. Interestingly, CDK19 tagged with HA, when expressed in flies, rescued defects associated with the loss of cdk8 in neurons, demonstrating functional conservation. Strikingly, the exogenous CDK19 protein was localized primarily in the cytoplasm of neurons, not the nucleus, suggesting that Cdk8/CDK19 may play an important role in the cytoplasm.
In this study, we show that depletion of Cdk8 in muscle and neurons causes a loss of mitochondrial fission, a phenotype that is also observed when Drp1, a protein required for mitochondrial fission, is lost11. These defects can be rescued by the expression of a cytoplasmically targeted CDK19 protein, indicating a non-nuclear role for Cdk8 family members. We endogenously tagged Cdk8 and found a dynamic subcellular distribution, including both cytoplasmic and nuclear localization in diverse tissues. We show that Cdk8 binds to Drp1, increases the phosphorylation of Drp1 at S616, and that loss of Cdk8 decreases the phosphorylation of Drp1 S616. Loss of Pink1, a kinase required for phosphorylation of Drp1 at S616, causes defects in mitochondrial dynamics, mitophagy, and Complex I function. Mutations in PINK1 and PARKIN have been identified in patients with juvenile Parkinsonism, a familial form of Parkinson’s Disease (PD)12,13,14,15. Pink1 is a serine/threonine kinase that regulates mitochondrial quality control16,17,18,19. pink1 mutant flies exhibit phenotypes resembling symptoms of PD patients, including defects in locomotor activity, reduced lifespan, and accumulation of dysfunctional mitochondria16,17,20,21,22,23. Overexpression of Cdk8 and both wildtype and cytoplasmic CDK19 rescues phenotypes associated with loss of pink1. In summary, our data reveal a new function for Cdk8/CDK19 in mitochondrial fission by mediating the phosphorylation of Drp1 and this function parallels the function of pink1.
Results
Depletion of Cdk8 causes altered wing posture, reduced lifespan, and defects in locomotion
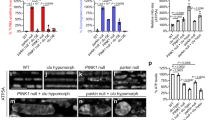
To investigate the consequences of reduced Cdk8 levels in flies, we examined the effects of ubiquitous knockdown of Cdk8 using the Act5c-Gal4 strain (Act5c>Cdk8 RNAi). Cdk8 expression was reduced by ~80% (Fig. S1a), and only about 50% of the flies eclosed at 25 °C (Fig. S1b). The flies that eclosed exhibit abnormal wing postures with either held-up or droopy wings shortly after eclosion (Fig. 1a). They also have significantly shorter life spans (Fig. S1c) and are sterile (Fig. S1d). We observed that Act5c>Cdk8 RNAi have impaired climbing abilities (Fig. 1b, c). Knocking down Cdk8 using either a muscle-specific (Mef2-Gal4) or a neuron-specific (Appl-Gal4) driver caused similar, though slightly less severe, climbing defects (Fig. 1b, c) when compared to ubiquitous knockdown. When a kinase-dead version of Cdk8 (Cdk8KD), which carries a single amino acid mutation within the well-conserved kinase domain (T173D, ref. 24), was expressed in muscles using Mef2-Gal4 it caused similar reduced survival (Fig. S1e) and severe climbing defects in escaper adults (Fig. S1f).
a Wing postures of flies expressing Act5c>RFP and Act5c>Cdk8 RNAi. b Violin plots showing distributions of climbing ability in control (RFP) and Cdk8 knockdown flies using either ubiquitous (Act5c), muscle (Mef2), or neuronal-specific (Appl) Gal4 drivers. c Average percent of flies that reached the target line of indicated genotypes in a climbing assay. Each experiment is individually repeated four times. The total number of progeny are assessed between white RNAi and Cdk8 RNAi with different drivers: n = 165, 144 in Act5>, 180, 189 in Mef2>, and 160, 160 in App>, respectively. Results are mean ± SD. An unpaired two-tailed t-test is used to generate the statistics. P value is 8.71e-07 for Act5c-Gal4, 1.67e-07 for Mef2-Gal4, and 2.32e-07 for Appl-Gal4. All raised at 25 °C. d–g Mitochondrial morphology of larval body wall muscles in (d) w1118 control or expressing (e) Cdk8 RNAi, (f) Cdk8WT, (g) Cdk8KD with Mef2-Gal4>mitoGFP, raised at 29 °C. Scale bar: 1 μm. d’–g’ Representative magnified sections from (d–g). Scale bar: 5 μm. h–j Quantification of mitochondrial morphology showing the (h) number of branches. P value is 0.0003 for Cdk8 RNAi, 0.4533 for Cdk8, and 6.97e-10 for Cdk8KD (i) the mean mitochondrial length. P value is 0.0023, 6.29e-8, and 0.0178, respectively, in order. j The mitochondrial area. P value is 0.0002, 0.5512, and 0.0167, respectively, in order, in larval body wall muscles of the indicated genotypes. All in comparison to w1118. Letters (d–g) refer to the genotypes shown in panels (d–g). Quantifications were calculated by the MiNA toolset. n = 6 per genotype. Data were presented as box plots (center line at the median, upper bound at 75th percentile, lower bound at 25th percentile) with whiskers at minimum and maximum values. An unpaired two-tailed t-test is used to generate the statistics. k–n Mitochondrial morphology of adult IFM in (k) w1118 control or expressing (l) Cdk8 RNAi, (m) Cdk8WT, (n) Cdk8KD with Mef2-Gal4>mitoGFP. Flies were grown at 29 °C, except escapers expressing Cdk8KD were raised at 25 °C. Scale bar: 10 μm. k’–n’ Representative magnified sections from (k–n). Scale bar: 5 μm. o–p Quantification of mitochondrial morphology showing the (h) number of mitochondria, p value is 0.0371 for Cdk8 RNAi, 4.25e-9 for Cdk8, and 0.0095 for Cdk8KD. i The mean mitochondrial length of the indicated genotypes. P value is 1.56e-8, 0.l0809, and 7.89e-9, respectively in order. All in comparison to w1118. n = 5 per genotype. Data were presented as box plots (center line at the median, upper bound at 75th percentile, lower bound at 25th percentile) with whiskers at minimum and maximum values. An unpaired two-tailed t-test is used to generate the statistics. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, ns no significance. Source data are provided as a Source Data file.
These phenotypes mimic phenotypes seen in flies with wing hinges and muscle defects25,26, including flies in which pink1 or parkin are depleted16,17,21,27. Mutations in PINK1 cause a familial form of Parkinson’s Disease27,28,29 and loss of pink1 disrupts mitochondrial homeostasis and impairs mitophagy23. Since the Cdk8 module is a well-known transcriptional regulator, we asked if the phenotypes were due to its role in regulating the transcription of pink1 and parkin using qRT-PCR. We found no significant differences in mRNA levels of pink1 or parkin when either Cdk8 or CycC were knocked down ubiquitously (Fig. S2a, b).
Cdk8 regulates mitochondrial morphology
Our previous data suggest that Cdk8/CDK19 may have a critical function in the cytoplasm. Given that most patients with de novo variants in CDK19 exhibit muscle hypotonia9 and that numerous genetic diseases that affect mitochondrial function cause hypotonia30, we decided to explore mitochondrial phenotypes in muscles. We further investigated the mitochondrial status following Cdk8 depletion, given the phenotypic similarity (Fig. 1a) to pink1 mutants that exhibit muscle defects. Mitochondria are highly dynamic and undergo fission and fusion31,32. We examined mitochondrial morphology in larval muscles using a muscle-specific driver and labeled mitochondria with mitoGFP33. In control larval body wall muscles (Mef2>mitoGFP/+), we observe punctate and filamentous structures (Fig. 1d). However, when Cdk8 levels are reduced using RNAi, we observe an increase in branch length, mitochondrial length, and total mitochondrial area (Fig. 1e, e’, h–j and Fig. S3b, c’). In contrast, overexpression of the wild-type Cdk8 cDNA does not affect branch number but causes a decrease in length (Fig. 1f, f’, h–j and Fig. S3d, e’). Moreover, overexpression of a kinase-dead version of Cdk8 (Cdk8KD) leads to very elongated mitochondria with significantly increased numbers of branches (Fig. 1g, g’ and Fig. S3f, f’). In summary, loss of Cdk8 or expression of kinase-dead Cdk8 affects mitochondrial dynamics in developing muscles.
We next examined mitochondrial morphology in adult indirect flight muscles (IFMs). In control animals, we observe both elongated and fragmented mitochondria (Fig. 1k, k’). Cdk8 depletion causes elongated mitochondria (Fig. 1l, l’), whereas overexpression of Cdk8 leads to small and round mitochondria, indicative of increased fission (Fig. 1m, m’). Since expression of Cdk8KD causes pupal lethality at 29 °C (Fig. S1e), we examined the mitochondrial morphology of escapers raised at 25 °C (Fig. 1n, n’) and again observed elongated mitochondria. These results were similarly observed when we tested the mitochondrial morphology using a different RNAi line for Cdk8 (Fig. S3b, c) or UAS-Cdk8-HA (Fig. S3i, j’). Since the Cdk8 module can modulate gene expression, we tested the effect of Cdk8 and Cyc C on the transcription of regulators of mitochondrial dynamics. Ubiquitous knockdown of either Cdk8 or Cyc C led to a modest increase in expression of both a regulator of mitochondrial fusion (mitochondrial assembly regulatory factor, marf) and fission (Drp1), suggesting the mitochondrial morphology defects are not likely due to reduced mRNA levels of these genes (Fig. S3n). Cyclin C (Cyc C) has previously been shown to exit the nucleus under oxidative stress conditions to regulate mitochondrial morphology in yeast and mouse embryonic fibroblast34,35,36. Depletion of Cyc C in muscle leads to elongation of mitochondria, and overexpression of Cyc C promotes fragmentation of mitochondria (Fig. S3g, h’). In summary, both Cdk8 and Cyc C modulate mitochondrial morphology, and proper expression of Cdk8 and Cyc C maintains the proper balance in mitochondrial dynamics. To test whether Cdk8 might function by promoting the activity of Cyc C, we tested whether Cyc C expression could rescue Cdk8 depletion phenotypes (Fig. S2c). While Cdk8 expression rescued the mitochondrial phenotypes caused by an expression of Cdk8-RNAi, Cyc C expression failed to rescue the phenotypes. We also performed the reciprocal experiment in which we tested whether Cdk8 could rescue the effects of Cyc C knockdown (Fig. S2d). Cdk8 expression alters mitochondrial abnormalities caused by an expression of Cyc C-RNAi but does not rescue. However, expression of Cyc C can rescue the fusion caused by the depletion of Cyc C. Hence, these results suggest that Cdk8 and Cyc C use distinct mechanisms to modulate mitochondrial dynamics.
Loss of Cdk8 alters mitochondrial distribution in the CNS neuropil
Given that patients with CDK19 variants develop severe defects in neurodevelopment, we next assessed the effects of depletion of Cdk8 in the nervous system. To assess mitochondrial distribution in the adult CNS, we expressed Cdk8 RNAi in neurons using a pan-neuronal driver, elav-Gal4, and stained the mitochondria with an anti-ATP5 antibody. As shown in Fig. 2a, mitochondria are indeed more clustered in cell bodies, and less abundant in the neuropil when compared to control animals (elav>control RNAi) (Fig. 2a). We further quantified the length of mitochondrial area and branches to assess the morphological changes. As shown in Fig. 2b, the area of mitochondria is significantly increased, but branch length is not altered (Fig. 2c).
a Mitochondrial distribution in the CNS neuropil. ATP5a marks mitochondria in the brain of control (elav>control RNAi) and flies that express Cdk8 RNAi (elav>Cdk8 RNAi). Scale bar: 10 μm. b Quantification of relative mitochondrial footprint (n = 6 for control, n = 5 for elav>Cdk8 RNAi). c Quantification of relative mitochondrial branch length (n = 6 for control, n = 5 for elav>Cdk8 RNAi). d Western blot and quantification of Hsp60 from the heads of control (elav>control RNAi) and flies that express Cdk8 RNAi (elav>Cdk8 RNAi) (n = 3 per genotype). p = 0.0013. e Quantification of ATP level in the heads of control (elav>control RNAi) and flies that express Cdk8 RNAi (elav>Cdk8 RNAi) (n = 6 per genotype). Statistical analyses are one-way ANOVA followed by a Tukey post hoc test. Results are mean ± SEM (∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, ns no significance). Source data are provided as a Source Data file.
As the loss of mitochondrial fission can lead to mitochondrial stress37 we quantified mitochondrial stress by measuring the level of the mitochondrial stress protein Hsp6038. As shown in Fig. 2d, mitochondrial stress is significantly increased in adult brains. Furthermore, the function of mitochondria is also affected as the ATP synthesis level is significantly reduced upon depletion of Cdk8 (Fig. 2e). Taken together, our data indicate that loss of Cdk8 affects mitochondrial dynamics, which results in a mitochondrial transport defect into the neuropil as well as a dysfunction of the mitochondria.
Abnormal mitochondrial morphology can elevate the production of reactive oxygen species (ROS). Therefore, we performed dihydroethidium (DHE) staining to assess ROS in adult brains. Adult brains of elav>cdk8 RNAi shows a 40% higher level of ROS when compared to control animals (elav>Luciferase RNAi) (Fig. S4a, b). Flies expressing human CDK19 in a Cdk8-depleted background decrease the level of ROS by 20% (Fig. S4a, b), showing that CDK19 can partially rescue the elevated ROS due to loss of Cdk8. Similarly, expression of Cdk8 RNAi in adult indirect flight muscles (Mef2>Cdk8 RNAi) significantly increased the level of ROS when compared to the control muscles (Mef2>white RNAi), however overexpression of fly Cdk8 does not significantly affect the level of ROS (Fig. S4c–g).
Loss of Cdk8 in photoreceptors affects mitochondrial size and distribution in photoreceptor terminals
To determine mitochondrial size and distribution in photoreceptors (PR) and their synapses, we performed TEM in the retina and lamina upon reduction of Cdk8 levels using RNAi driven by Rhodopsin1-Gal4 (Rh1> Cdk8 RNAi). We found that the size of mitochondria is significantly reduced, and the number of mitochondria is significantly increased in the PR cell body, compared to control (Rh1 > UAS-Luciferase RNAi) and Cdk8 knockout flies rescued by expression of wild-type human CDK19 (Rh1>cdk8 RNAi; UAS-CDK19WT) (Fig. 3a, c). In addition, the number of mitochondria is reduced in photoreceptor terminals in Rh1>Cdk8 RNAi flies when compared to control flies or flies expressing wild-type human CDK19 in the Cdk8 RNAi background (Fig. 3b). Loss of Drp1 in photoreceptors leads to an increase in small mitochondria in the cell body of PR that are highly clustered as well as a loss of mitochondria at synapses39. The data suggested that loss of Drp1 leads mitochondria to be clustered like beads on a string and hence interconnected, and that they form a network that is poorly transported to synapses. These data clearly indicate that Cdk8 loss partially phenocopies the Drp1 loss phenotypes in photoreceptors. Hence, the loss of Cdk8 impairs mitochondrial fission in PR. Furthermore, these defects in mitochondria can be rescued by expression of wild-type human CDK19.
a Mitochondrial distribution in the cell bodies of the photoreceptors of the indicated genotypes. Scale bars: 2 μm. b TEM images of photoreceptor terminals. The size of mitochondria is smaller than in the control but observed more mitochondria in clusters in photoreceptors of flies that express Cdk8 RNAi (Left, Rh1>Cdk8 RNAi; UAS-mCherry) when compared to the control (Middle, Rh1>Cdk8 RNAi; UAS-CDK19WT). Scale bars: 2 μm. c Quantification of mitochondrial size and number in photoreceptors (n > 50 per genotype for the quantification of mitochondria size, n = 7 per genotype for the quantification of mitochondrial number). Quantification of mitochondrial number per photoreceptor terminal (Right) (n = 10 per genotype). Statistical analyses are one-way ANOVA followed by a Tukey post hoc test. Results are mean ± SEM (∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, ns no significance). Source data are provided as a Source Data file.
Given that there is a depletion of mitochondria at synapses as well as a reduction in ATP production, we surmised that there may be a defect in synaptic transmission. Indeed, ref. 39 argued that the recycling of synaptic vesicles at synapses requires the presence of functional mitochondria and proper ATP levels to drive the recycling process40. We therefore performed Electroretinograms (ERGs) in flies in which Cdk8 levels are reduced (elav>Cdk8 RNAi) and observed a dramatic reduction of on-and-off transients, indicating that the PR fails to communicate with the postsynaptic cells and that synaptic transmission is nearly abolished (Fig. S5a). In contrast, the ERG amplitudes are not altered, suggesting that the phototransduction cascade is not affected (Fig. S5b). These data are strikingly similar to the loss of Drp1 in photoreceptors39.
Cdk8 and CDK19 have context-dependent subcellular localization
We have previously shown that human CDK19WT was primarily localized to cytoplasm in neurons9. We thus sought to characterize the localization of CDK19 in several Drosophila tissues. Using an antibody to detect human CDK19, we confirmed the cytoplasmic localization in the adult central nervous system (Fig. 4a), while we observed nuclear localization in larval wing imaginal disks and salivary glands (Fig. 4a). In adult indirect flight muscles, CDK19WT was found in both cytoplasm and nuclei (Fig. 4c).
a Subcellular localization of UAS-CDK19WT-3xHA in adult CNS. b Subcellular localization of UAS-CDK19WT-3xHA and UAS-CDK19ΔNLS-3xHA in wing imaginal disks and salivary glands. Green indicates Dlg staining, which marks the cell membrane, and red indicates HA-tagged proteins. The stainings were independently repeated three times. Scale bar: 10 μm. c Subcellular localization of UAS-CDK19WT-3xHA and UAS-CDK19ΔNLS-3xHA in adult indirect flight muscles. Anti-HA is used to visualize CDK19 localization. DAPI was used to detect nuclear DNA and anti-lamin antibody highlights the nuclear membrane. Cross sections of confocal Z-stacks are shown in the right most column. d Myosin heavy chain (Mhc)-Gal4 was used to express Cdk83xHA in muscles with either control white RNAi or an additional copy of Cdk83xHA. Whole cell lysate (W), nuclear (N), and cytoplasmic (C) fractions were prepared, and antibodies to ATP5a and Histone 3 were used to detect the cytoplasmic and nuclear fractions, respectively. e Myc-tagged Cdk8 (magenta) localization in S2R+ cells. Co-staining with DAPI and ATP5a highlights cytoplasmic localization. Whole cell lysate (WCL), nuclear, and cytoplasmic fractions were blotted to detect Cdk8. Experiments were repeated three times. Source data are provided as a Source Data file.
These findings suggested that CDK19 may play important cytoplasmic roles. To assess the importance of the predicted nuclear localization sequence (NLS) (Fig. S6a, b), we generated UAS-CDK19 NLS transgenic flies with and without a 3XHA tag, respectively (UAS-CDK19ΔNLS-3XHA and UAS-CDK19ΔNLS). Phenotypes produced by both constructs were the same, showing that the 3XHA tag did not affect function. Expression of the tagged construct in wing disks and salivary glands using da-Gal4 shows that deletion of the NLS leads to a cytoplasmic localization (Fig. 4b). Expression of CDK19ΔNLS in indirect flight muscles showed only cytoplasmic localization. Co-staining with DAPI to detect nuclear DNA and anti-lamin antibodies, which detect the lamins in the nuclear membrane, highlights the exclusion from the nucleus (Fig. 4c). These findings validate that the predicted NLS sequences can target CDK19 to the nucleus.
Given the lack of available antibodies to detect endogenous Cdk8 in tissue, we relied on the HA tags encoded in the Cdk8 transgene to track the localization in subcellular fractions. Mhc>Cdk83xHA flies were crossed to either white RNAi or UAS-Cdk83xHA to produce flies expressing one or two copies of exogenous Cdk8. Nuclear and cytoplasmic fractions were isolated from whole cell lysates, and the presence of Cdk8 was detected with anti-HA antibodies (Fig. 4d). Antibodies to ATP5a were used to highlight the cytoplasmic fraction, while Histone 3 marked nuclear fractions. Cdk8 was found in both nuclear and cytoplasmic fractions, and the relative distribution was unchanged when Cdk8 was elevated. Interestingly, a recent study showed that CDK8 is primarily localized in cytoplasmic in murine pancreatic cells where it regulates insulin secretion41, which is in line with our observation on Cdk8.
Finally, to validate our observation, we further expressed Myc-tagged Cdk8 in Drosophila S2R+ cells, which are derived from late-stage embryos42, and observed a clear cytoplasmic localization (Fig. 4e, left panel). This distribution was confirmed by western blotting of nuclear and cytoplasmic fractions (Fig. 4e, right panel).
Endogenous GFP-tagged Cdk8 is found in nuclei and the cytoplasm
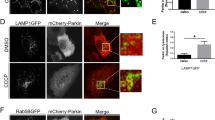
To gain more insight into endogenous Cdk8 localization, we used a CRISPR-based approach to generate a Cdk8-superfolder-GFP fusion protein fly line (referred to as Cdk8::GFP) in which the resulting endogenous Cdk8 protein is tagged with superfolder GFP (Fig. 5a)43. The construct also encodes a scarless-DsRed in the untranslated region of Cdk8, which aids in the selection of transformed flies. Cdk8::GFP homozygous flies are viable and do not exhibit any overt phenotypes, indicating that insertion of sfGFP does not affect Cdk8 function. Through this GFP insertion, the detection of GFP reflects the endogenous Cdk8 protein expression and localization. In these experiments, we detected the GFP fluorescence without the use of anti-GFP antibodies, to eliminate any chance of non-specific staining. In larval body wall muscles, Cdk8::GFP is primarily nuclear, though low-level cytoplasmic staining is seen throughout the tissue when compared to the control (Fig. 5b). In adult brains, Cdk8 shows a dynamic distribution (Fig. 5c) characterized by nuclear and cytoplasmic staining (Fig. 5d). Some cells show primarily nuclear while others show primarily cytoplasmic localization (Fig. 5e), highlighting the variability in Cdk8 protein localization and demonstrating in vivo that endogenous Cdk8 is present in the cytoplasm. We used fractionation of adult brain protein lysates to further validate the cytoplasmic localization of Cdk8::GFP (Fig. S7a).
a Schematic illustration for the generation of endogenously tagged Cdk8::GFP flies. b Subcellular localization of Cdk8::GFP in larval body wall muscles in comparison to w1118, as well as in the adult brain. c Positive signals rely on the endogenously expressed GFP. n = 10 per genotype. d Single plane, zoom-in images with orthogonal views of adult brains comparing w1118 and Cdk8::GFP. n = 10 per genotype. The selected region is shown in (e) to further demonstrate the cytoplasmic localization of endogenous Cdk8.
Cdk8 interacts with Drp1 and promotes its phosphorylation and mitochondrial fragmentation
Given that the variants observed in human CDK19 patients are enriched in the kinase domain9 and since Cdk8KD acts as the dominant negative variant when expressed in flies, we assume that the kinase function of Cdk8 may regulate mitochondrial dynamics. We performed protein sequence alignments for the CDK consensus phosphorylation motif S/T-P-X-R/K44 for regulators of mitochondrial dynamics, including Opa1, Marf, and Drp145. We did not find the motif in the fusion proteins (Opa1and Marf), but the fission regulator Drp1 carries the motif at serine 616 (Fig. S5c), a site that is known to be regulated by phosphorylation, acetylation, and SUMOylation46. These findings suggest that the effects of mitochondrial dynamics of Cdk8 may be mediated by the fission regulator, Drp1.
To test if endogenous GFP-tagged Cdk8 can physically interact with Drp1, we performed co-immunoprecipitation using fly muscles expressing HA-tagged Drp1 in the Cdk8::GFP fly strain (Fig. 6a). Drp1 was pulled down using anti-HA beads and Cdk8::GFP was detected using an anti-GFP antibody in co-immunoprecipitations of Drp1 but not with anti-IgG agarose. This finding shows that Drp1, which is cytoplasmic, can interact with endogenous GFP-tagged Cdk8. We also performed co-immunoprecipitation using S2R+ insect cells co-transfected with Myc-tagged Cdk8 and HA-tagged Drp1 plasmids (Fig. S7b). Drp1 was pulled down with Cdk8 in the Myc-tagged Cdk8 immunoprecipitation (IP), while Drp1 was absent from the control IgG IP. Cdk8 was also pulled down in the reciprocal pulldown using HA-tagged Drp1 (Fig. S7b). These findings show that Cdk8 physically interacts with Drp1.
a Co-immunoprecipitation using adult thoraxes expressing MhcGal4 > UAS-Drp1HA+Cdk8::GFP at 25 °C. IgG agarose beads were used as a negative control. IPs used HA-agarose beads and detected endogenous Cdk8::GFP using rabbit anti-GFP antibody. Experiments were individually repeated three times. b Western blot showing the level of total Drp1 or S616 phospho-Drp1 in control (Mef2>white RNAi), Mef2>Cdk8-RNAi and Mef2>Cdk8HAflies. Quantifications of blots normalized to actin levels (n = 3 per genotype). Results are mean ± SD. An unpaired two-tailed t-test is used to generate the statistics. P value is 0.1850 for Cdk8 RNAi, and 0.4490 for Cdk8 in comparison to white RNAi. Flies were raised at 25 °C. c Myc-Cdk8 and HA-Drp1 from S2R+cytoplasmic fractions were immunoprecipitated with Myc-agarose and subjected to in vitro kinase assays in the presence or absence of ATP. Experiments are repeated three times. Results are mean ± SD. An unpaired two-tailed t-test is used to generate the statistics. P value is 0.0149 for Drp1 and 8.54e-05 for pDrp1/Drp1. Samples were probed with antibodies to Myc to quantify Myc-Cdk8 levels (which were equivalent between conditions), Drp1 to detect the degree of Drp1 binding to Cdk8, and with anti-pDrp1S616 to quantify the degree of phosphorylation relative to total Drp1. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, ns no significance. Source data are provided as a Source Data file.
We next asked if Cdk8 expression affects the abundance of Drp1 or S616-phosphorylated Drp1. We examined the levels of both endogenous total Drp1 and phospho-S616-Drp1 in lysates from fly tissues in which Cdk8 was either knocked down or overexpressed in larval body wall muscles. We found no significant difference in the level of Drp1 protein between the control and either Cdk8-RNAi or Cdk8 overexpression (Fig. 6b). However, an antibody that recognizes pDrp1S616, shows a significant decrease following Cdk8 depletion and an increase in pDrp1S616 when Cdk8 is expressed (Fig. 6b). To determine if loss of Cdk8 affects the phosphorylation of Drp1 in neurons, we assessed the protein levels of phospho-Drp1 S616 and normal Drp1 in adult heads by performing Western blot using the extracts of heads of elav>Cdk8 RNAi flies. We found that the phospho-Drp1 S616 level is decreased by about 25% (Fig. S5d). In summary, the data show that Cdk8 interacts with Drp1 and promotes phosphorylation of Drp1 at S616, and activates the mitochondrial fragmentation process. Our data strongly suggest that the effect of Cdk8 on Drp1 is direct, although we cannot rule out entirely that it is indirect via an associated kinase.
Given that Cdk8 and Drp1 interact and are both cytoplasmic, we immunoprecipitated Myc-Cdk8 from the cytoplasmic fractions of S2R+ cells transfected with both Myc-Cdk8 and HA-Drp1 constructs. The immunoprecipitated proteins were then subjected to in vitro kinase assays in the absence or presence of ATP (Fig. 6c). We quantified the total amount of Cdk8, which was equivalent between experimental conditions. We next determined relative amounts of total Drp1 and pS616-Drp1. We found that phosphorylation of co-immunoprecipitated cytoplasmic Drp1 is significantly elevated in the presence of ATP (Fig. 6c).
Cytoplasmic CDK19 replaces the function of Cdk8 in neurons and rescues mitochondrial defects
The data presented so far in muscles and neurons argue that Cdk8 and CDK19 function in the cytoplasm to promote mitochondrial fission by phosphorylating Drp1. However, in muscles as well as in the developing third instar larval brains Cdk8 is present in nuclei and the cytoplasm, begging the question of whether the cytoplasmic localization is sufficient for this function9. To determine if an expression of cytoplasmic CDK19 can rescue the loss of Cdk8 in the nervous system, we expressed Cdk8 RNAi in neurons (elav>Cdk8 RNAi) and examined the effects of expressing either the reference CDK19 (UAS-CDK19WT) or UAS-CDK19ΔNLS. As shown in Fig.7a, expression of CDK19ΔNLS is sufficient to rescue the lifespan defects observed in flies expressing Cdk8 RNAi in neurons (elav>Cdk8 RNAi) similar to CDK19WT expression (Fig. 7a).
a CDK19 ΔNLS can suppress the lifespan decrease caused by an expression of Cdk8 RNAi in the neuron to a similar extent to CDK19WT expression (n > 100 per genotype). b Quantification of acridine orange staining (AO), which assesses neuronal death. when Cdk8 is decreased in adult brain neurons. Red indicates the dying neurons. (n = 7, n = 14, n = 8, and n = 16 respectively in order). Scale bar: 10 μm. Statistical analyses are one-way ANOVA followed by a Tukey post hoc test. Results are mean ± SEM (∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, ns no significance). c Both CDK19WT and CDK19 ΔNLS can significantly suppress the climbing defects due to neuronal depletion of cdk8 (elav>Cdk8 RNAi). Experiments are repeated four times. Results are mean ± SD. An unpaired two-tailed t-test is used to generate the statistics. P value is 4.56e-05 for white RNAi, 2,38e-05 for CDK19WT, and 0.0036 for CDK19ΔNLS in comparison to Cdk8 RNAi. The total number of progeny are assessed, n = 160, 153, 140, 159, respectively, in order. d–g Rhodamine-phalloidin staining (F-actin, red) to visualize indirect flight muscles in (d) control white RNAi or (e) Cdk8 RNAi flies. f CDK19WT and (g) CDK19ΔNLS were expressed in the Cdk8-depleted background. d’–g’ Either mitoGFP or ATP5α staining (magenta) were used to visualize mitochondrial morphology in the indicated genotypes. Scale bars: 10 μm. h Quantification of adult indirect flight muscle width in 3-day-old flies. n = 5 per genotype. Data were presented as box plots (center line at the median, upper bound at 75th percentile, lower bound at 25th percentile) with whiskers at minimum and maximum values. An unpaired two-tailed t-test is used to generate the statistics. P value is 1.78e-16 for white RNAi, 0.2295 for CDK19WT, and 0.1816 for CDK19ΔNLS in comparison to Cdk8 RNAi. All raised at 25 °C. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, ns no significance. Source data are provided as a Source Data file.
We next assessed cell death induced by elav>Cdk8 RNAi in the brain in 10-day old flies kept at 25 °C by staining with acridine orange (AO) and quantified fluorescence intensity. We observed that an elevated level of AO can be significantly reduced by expression of either UAS-CDK19WT or UAS-CDK19ΔNLS (Fig. 7b). Depletion of Cdk8 in neurons leads to impaired climbing ability (Fig. 1b) which can also be rescued by expression of either UAS-CDK19WT or UAS-CDK19∆NLS (Fig. 7c). These findings argue strongly that the function of Cdk8 in these assays is cytoplasmic.
We also examined the effects of wildtype and cytoplasmic CDK19 in muscles. To visualize mitochondrial morphology, we used either mitoGFP or an antibody against ATP5α (Fig. 7d’–g’), whereas muscle fibers were visualized using rhodamine-phalloidin to detect filamentous actin (Fig. 7d–g). Cdk8 knockdown leads to a widening of muscle fibers compared to control (Fig. 7d–h). Expression of either UAS-CDK19WT or UAS-CDK19∆NLS restored muscle fiber morphology equally well (Fig. 7f–h). We next examined mitochondrial morphology. We observed that expression of both UAS-CDK19WT or UAS-CDK19∆NLS in a wild-type background led to rounded and fragmented mitochondria, indicating they can promote fission (Fig. S3l’, m’). A mixture of elongated and fragmented mitochondria was found in control (Fig. 7d), and depletion of Cdk8 caused a highly elongated mitochondrial morphology (Fig. 7e). We observed predominantly fragmented mitochondria when either UAS-CDK19WT or UAS-CDK19∆NLS was co-expressed in Mef2>Cdk8 RNAi background (Fig. 7f’, g’). These results show that cytoplasmic CDK19 is sufficient to regulate mitochondrial morphology.
Expression of Cdk8 can rescue pink1 muscle degeneration and locomotor defects
Our results so far show that Cdk8 regulates mitochondrial fission by phosphorylating Drp1. Interestingly, Pink1 was previously shown to directly phosphorylate Drp1 at S616 to promote its activity and fragmentation of mitochondria47,48. Given that both Cdk8 and Pink1 phosphorylate the same amino acids of Drp1 and given that they are both required for normal mitochondrial function neither alone may be sufficient to fully phosphorylate Drp1 at S616. Hence, overexpression of Cdk8 may be able to suppress the phenotypes associated with the pink1 nulls (pink1B9), a fly model for PD17. pink1 is on the X chromosome and hemizygous pink1B9 males were used in subsequent experiments. Loss of pink1 causes thorax indentation (Fig. 8a), an established indicator for the underlying muscle degeneration17. Muscle degeneration is prominent in pink1 or parkin mutant flies where apoptotic cell death leads to thorax indentation and flight and mobility defects17,21. Approximately 90% of pink1B9 flies have thorax indentation, and expression of human Pink1 (hPink1) using the muscle driver, Mef2-Gal4, fully rescues the phenotype (Fig. 8a). Mef2>Cdk8 expression also significantly rescued the phenotype as only 40% of flies showed thorax indentation defects (Fig. 8a).
a Thorax indentation (white arrowhead indicates indentation) with quantification of percent indentation in 3-day-old flies expressing either RFP, hPink1, or Cdk8 with Mef2-Gal4 in the pink1B9 mutant background compared to FM7/Y control flies. Experiments are repeated four times. The total number of progeny are assessed, n = 120, 130, 137, 119, respectively, in order. Results are mean ± SD. An unpaired two-tailed t-test is used to generate the statistics. P value is 1.30e-6 for both FM7/Y and hPINK1 and 0.01 for Cdk8 in comparison to the pink1B9 mutant. b Violin plots showing the distribution of climbing activity of RFP, hPink1, or Cdk8 with Mef2-Gal4 in the pink1B9 mutants. The average percent of flies that reach the target line in indicated genotypes. Experiments are repeated three times. The total number of progeny are assessed, n = 80, 74, 65, respectively, in order. Results are mean ± SD. An unpaired two-tailed t-test is used to generate the statistics. P value is 3.96e-6 for hPINK1 and 6.62e-5 for Cdk8 in comparison to the pink1B9 mutant. Raised at 29 °C. c Climbing assay testing the ability of RFP, CDK19WT, or CDK19∆NLS expressed in the pink1B9 background to reach the target line. Experiments are repeated three times. The total number of progeny are assessed, n = 56, 71, 98, respectively, in order. Results are mean ± SD. An unpaired two-tailed t-test is used to generate the statistics. P value is 0.0004 for CDK19WT and 0.0002 for CDK19 ΔNLS in comparison to the pink1B9 mutant. d Quantification of thorax indentation in flies when RFP, CDK19WT, or CDK19ΔNLS are expressed in the pink1B9 background, raised at 25 °C. Experiments are repeated three times. Total number of progeny are assessed, n = 56, 77, 84. Results are mean ± SD. An unpaired two-tailed t-test is used to generate the statistics. P value is 0.0025 for CDK19WT and 0.0238 for CDK19 ΔNLS in comparison to the pink1B9 mutant. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Source data are provided as a Source Data file.
Next, we tested the effect of Cdk8 expression on the severe locomotor defects observed in pink1B9 flies (Fig. 8b). pink1B9 climbing defects were significantly rescued by expression of either hPINK1 or Cdk8 in 3-day old flies (Fig. 8b). Thorax indentation (Fig. S8a) as well as climbing ability (Fig. S8b) were also examined in 3-week-old flies. Both hPINK1 and Cdk8 significantly rescued these phenotypes in these aged flies. In summary, Cdk8 expression can significantly rescue thorax indentation and climbing defects in pink1B9 mutants.
Cytoplasmic CDK19 can rescue pink1 B9 phenotypes
We observed that either overexpression of CDK19WT or CDK19∆NLS is sufficient to induce severe mitochondrial fragmentation, as observed with the expression of Cdk8. These results led us to wonder if their expression could rescue the pink1B9 mutant phenotypes, similar to the expression of Cdk8. We assayed the climbing activity (Fig. 8c) and thorax indentation (Fig. 8d) phenotypes when either CDK19WT or CDK19∆NLS were expressed in a pink1B9 mutant background. Strikingly, both wildtype and cytoplasmic CDK19∆NLS significantly rescued both phenotypes (Fig. 8c, d). These findings show that the functions of Cdk8 are conserved in cytoplasmic CDK19.
Cdk8 expression rescues mitochondrial defects and muscle degeneration in pink1 B9 flies
We next examined the muscle and mitochondrial morphology in dorsal longitudinal adult indirect flight muscles in the thorax49,50. We used mitoGFP to detect mitochondria and rhodamine-phalloidin to visualize F-actin and overall muscle integrity in dissected IFMs. In control male flies (FM7/Y), elongated and tubular mitochondria (Fig. 9a) are found evenly distributed and intercalated between well-defined muscle fibers (Fig. 9a”). Mitochondrial morphology was severely disrupted in pink1B9 mutants as previously reported (Fig. 9b)16,17,21: they exhibit a reduced number of mitochondria, irregularly shaped mitochondria that are scattered between the muscle fibers, and the muscle fibers are significantly narrower in width than in control animals (Fig. 9b’)16,17,20,21,23,51. Aggregates of mitochondria are also seen (white arrowheads in Fig. 9b)52,53. Expression of both hPINK1 and Cdk8 rescued the defects in both mitochondria and muscle (Fig. 9c, d”). Myofibril width quantification revealed that expression of either hPINK1 or Cdk8 in pink1B9 mutants significantly suppressed muscle degeneration (Fig. 9e). Together, these findings show that elevated expression of Cdk8 can suppress mitochondrial and muscle defects observed in the pink1B9 PD model.
a–d Mitochondrial morphology in indirect flight muscle (IFM) in (a) FM7/Y, (b) pink1B9/Y, and either (c) hPINK1, or (d) Cdk8 expressed in the pink1B9 mutant background with Mef2>mitoGFP. a′–d′ Rhodamine-phalloidin staining (magenta) stains actin filaments to visualize the muscle morphology. a″–d″ Merged images, including DAPI staining for DNA (blue). Scale bars: 10 μm. e Quantification of adult indirect flight muscle width in 3-day-old flies (n = 5 per genotype). Data were presented as box plots (center line at the median, upper bound at 75th percentile, lower bound at 25th percentile) with whiskers at minimum and maximum values. An unpaired two-tailed t-test is used to generate the statistics. P value is 1.13e-024 for FM7/Y, 1.75e-23 for hPINK1, and 1.42e-22 for Cdk8 in comparison to the pink1B9 mutant. f–i IFM in (f) FM7/Y, (g) pink1B9, and (h) hPink1 or (i) Cdk8 expressed in the pink1B9 mutant background with Mef2>mitoGFP raised at 29 °C. f′–i′ DHE staining of corresponding genotypes in (f–i). Scale bars: 100 μm. j Quantification of mean DHE fluorescence intensity in live adult IFM of the indicated genotypes (n = 8 per genotype). Data were presented as box plots (center line at the median, upper bound at 75th percentile, lower bound at 25th percentile) with whiskers at minimum and maximum values. An unpaired two-tailed t-test is used to generate the statistics. P value is 3.11e-25 for FM7/Y, 1.71e-26 for hPINK1, and 1.46e-12 for Cdk8 in comparison to the pink1B9 mutant. All raised at 29 °C. k Western blot showing the level of total Drp1 or S616 phospho-Drp1 in control (Mef2>white RNAi), pink1 mutant flies (pink1B9; Mef2>white RNAi) and pink1 expressing Cdk8 (pink1B9; Mef2>Cdk83xHA. Quantifications of blots normalized to actin levels. (n = 3 per genotype). Results are mean ± SD. An unpaired two-tailed t-test is used to generate the statistics. P value is 0.0039 for FM7 and 0.0424 for Cdk8 in comparison to pink1B9 mutant for detecting the level of pDrp1S616. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, ns no significance. Source data are provided as a Source Data file.
Cdk8 suppresses ROS production in pink1 B9 mutants
Loss of Pink1 causes accumulation of dysfunctional mitochondria, which impairs the electron transport chain (ETC) and, in turn, leads to elevated production of reactive oxygen species (ROS)54. Since an expression of Cdk8 can alleviate the mitochondrial phenotypes in pink1B9, we asked if the morphologically restored mitochondria are also functionally restored. We used dihydroethidium (DHE) staining to assess ROS in adult indirect flight muscles55. The FM7/Y control males show redox homeostasis under physiological conditions (Fig. 9f, f’). In contrast, pink1B9 flies show enhanced DHE intensity, indicative of elevated ROS production56,57 (Fig. 9g, g’), and elevated DHE staining is observed near aggregated mitochondria (Fig. S8d, d”, yellow arrowhead). hPINK1 significantly reduced mitochondrial aggregation and suppressed ROS elevation (Fig. 9h and S8e, e’). ROS production in Cdk8-expressing pink1B9 flies was also reduced, and mitochondrial integrity was restored (Fig. 9h’ and S8f-f”). Quantification of DHE mean intensity demonstrated that either hPINK1 or Cdk8 significantly suppressed the elevated ROS levels caused by pink1B9 mutants (Fig. 9j). We also used a spectrophotometer to measure emitted DHE fluorescence from adult thoraxes (Fig. S8g), confirming that elevated ROS found in pink1B9 flies is significantly suppressed by expression of either hPINK1 or Cdk8.
Cdk8 rescues pink1-induced decreases in phospho-Drp1
Our results suggest that Cdk8 can promote Drp1 activity through phosphorylation and mitochondrial fission. Since Pink1 can also carry out this role, we asked whether, in the absence of functional Pink1, elevated expression of Cdk8 could restore Drp1 phosphorylation levels. Using fly lysates from control flies, pink1 mutants, and pink1 expressing wild-type Cdk8, we found that the overall levels of endogenous Drp1 were similar in all three samples (Fig. 9k). In contrast, pink1 mutants showed significant decreases in pDrp1S616, relative to control, and these decreased levels were significantly rescued by expression of Cdk8 (Fig. 9k).
Discussion
Here we show that depletion of Cdk8 in muscle causes a severe reduction of mitochondrial fission, a phenotype that is also observed when Drp1 is lost. We show that Cdk8 binds to Drp1, phosphorylates Drp1 at S616, and that loss of Cdk8 leads to decreased phosphorylation of Drp1 S616 in muscles and neurons. Interestingly, pink1 also phosphorylates Drp1 at S616, and loss of pink1 causes defects in mitochondrial dynamics, mitophagy, and Complex I function. Overexpression of Cdk8 rescues phenotypes associated with loss of pink1, including locomotor impairment and defects in mitochondrial morphology and muscle integrity. The data indicate that Cdk8 functions similarly to Pink1. We demonstrate that endogenous Cdk8 can localize to the cytoplasm in muscles and brain supporting our model that Cdk8 has cytoplasmic functions. In summary, our data reveal a new cytosolic function for Cdk8/CDK19 in mitochondrial fission by mediating the phosphorylation of Drp1 and this function parallels the function of pink1.
Functional conservation of Cdk8 and CDK19
CDK8 and its paralog, CDK19, are cyclin-dependent kinases known to regulate transcription. Either CDK8 or CDK19 are components of a 4-protein complex called the Mediator kinase module1. However, they are mutually exclusive in this complex. This module reversibly associates with Mediator, a 26-subunit protein complex that regulates RNA Polymerase II-mediated gene expression. As part of this complex, the other Mediator kinases have been implicated in diverse processes such as developmental signaling, metabolic homeostasis, and innate immunity58. In recent years, dysregulation of Mediator kinase module proteins, including CDK8/19, have been implicated in the development of different human diseases, in particular, cancer59.
In mice and humans, CDK8 is expressed ubiquitously, whereas CDK19 is enriched in the brain60. Loss of CDK8 results in embryonic lethality in mice61, whereas loss of CDK19 causes a mild phenotype (International Mouse Phenotyping Consortium; https://www.mousephenotype.org/). The latter mice are born at expected Mendelian ratios, and the only obvious phenotype is an increase in grip strength. In contrast, in humans, variants in CDK19 act as dominant negative variants and cause developmental delay, infantile spams, hypotonia, and intellectual disability9. Several CDK8 variants located in the kinase domain also behave as dominant negative alleles and cause hypotonia, facial dysmorphisms, and congenital heart disease7,8. Thus far, all phenotypes associated with CDK8 and CDK19 are thought to be related to their nuclear function.
We previously observed that human CDK19 is localized to the cytoplasm in adult fly neurons9. Moreover, flies that have reduced Cdk8 protein in most cells displayed phenotypes that were strikingly similar to the ones observed in pink1 null mutants17,62. Importantly, overexpression of Cdk8 significantly suppresses the behavioral and mitochondrial defects associated with the loss of pink1, which encodes a putative serine/threonine kinase with a mitochondrial targeting sequence. Pink1 promotes mitochondrial clearance, and phosphorylates Drp1 at site S616 in mice and human fibroblast47. Hence, these data implicate CDK19/Cdk8 in a critical mitochondrial function.
Regulation of Drp1 by Cdk8/CDK19 and PINK1
Perturbations in mitochondrial morphology have been associated with several human disorders63. Some of the genes are involved in regulating fission and fusion64. Mitofusins (Mfn1 and Mfn2) regulate mitochondrial outer membrane fusion and are required to maintain a mitochondrial network in cells, while mitochondrial inner membrane protein Opa1 regulates mitochondrial inner membrane fusion65. Drp1, a cytosolic dynamin GTPase, is the key player in mitochondrial fission. Importantly, Drp1 phosphorylation plays a critical role in controlling mitochondrial fission66. Phosphorylation of Drp1 at the S616, S579, and S600 sites promotes fission during mitosis and oxidative stress67,68,69,70. We show that Cdk8 upregulates the phosphorylation of Drp1 at S616 and hence acts in a very similar fashion as Pink1. This observation, combined with the fact that overexpression of Cdk8 nearly fully suppresses climbing and mitochondrial phenotypes associated with loss of pink1 strongly argues that Cdk8 plays a critical and similar role in mitochondrial dynamics as Pink1. Hence, it is likely that another downstream effector of Pink1, Parkin, may also be affected by Cdk8/CDK19. However, loss of pink1 also affects iron-sulfur cluster synthesis, which leads to a cascade of events that are independent of mitophagy, and these does not require Parkin62.
Drp1 and human diseases
DNM1L (Dynamin 1-like, synonyms Drp1) is a human homolog of Drp1, which is a member of the dynamin superfamily of GTPases. DNM1L mediates mitochondrial as well as peroxisomal fission71, and a patient with loss of DNM1L presented with a lethal encephalopathy due to defective mitochondrial and peroxisomal fission (MIM #614388)72. This individual had poor feeding, poor growth, lactic acidosis, seizures, hypotonia, nystagmus, and an abnormal gyral pattern on magnetic resonance imaging and passed away at 37 days of life71. Mice lacking Dlp1, the homolog of DNM1L, die at embryonic day E12.5, indicating a crucial role for this gene in mammalian development. These mice display abnormal mitochondrial and peroxisomal morphology73, indicating that Drp1 is critical in development across the species. The human CDK19 patients show some similar but milder phenotypes, including developmental delay, seizure, and hypotonia, suggesting that the function of DNM1L is not fully dependent on Pink1 and CDK19.
Mitochondrial dynamics under physiological conditions associated with the translocation of CycC and CDK19
Cyc C is a binding partner of CDK8/CDK19. Under normal conditions, CDK19-Cyc C regulates transcription through association with the Mediator complex of RNA polymerase II2. However, in contrast to other cyclins, the concentration of Cyc C does not change during cell-cycle progression74,75,76. Instead, previous work has shown that oxidative stress can induce a nuclear release of Cyc C to the mitochondria, where it facilitates fission through Drp1 and Mff in mouse embryonic fibroblasts35. Here we show that expression of Cdk8/CDK19 can modulate mitochondrial morphology under physiological conditions, likely through regulating Drp1, in a similar fashion as Cyc C. We found that the effects of Cdk8 depletion could not be rescued by ectopic Cyc C, suggesting that under our experimental conditions, the two proteins may act on Drp1 through distinct mechanisms.
Roles of CDK8/19 in rare diseases and implications for common disorders like Parkinson’s Disease
Patients harboring missense mutations in either CDK8 or CDK19 present with a range of clinical features, including neurodevelopmental defects, variable intellectual disability, hypotonia, and facial dysmorphology7,8,9,10. Thus far, the features of these rare syndromes have been attributed to dysfunction of these kinases as part of the Mediator kinase module. We propose that aspects of these syndromes may also be due to altered function in the cytoplasm in the regulation of mitochondrial morphology and function through effects on Drp1, independent of their role in mediating transcription. In this study, we show that loss of Cdk8 causes defects that show some similarity to Parkinsonism and both Cdk8 and CDK19 can suppress the phenotypes associated with fly pink1 mutants. These findings suggest CDK8/19 may also contribute to aspects of the development of Parkinson’s disease.
Methods
Drosophila culture
Drosophila melanogaster flies were raised on standard cornmeal-molasses food. Stocks were kept at 25 °C. Crosses were carried out at 25 and 29 °C as indicated. Since Gal4 activity is elevated at higher temperatures77, we could modulate expression levels using both temperatures. Results found at 29 °C were more penetrant, so the main figures include these data, and supplemental figures show data from 25 °C. The following fly strains were used: Act5c-Gal4/TM6B (BL #3954), Mef2-Gal4 (BL #27390), Appl-Gal4 (BL #32040), pink1B9 (BL #34749), UAS-Cdk8-RNAi (BL #35324), UAS-RFP (BL #7119), UAS-white-RNAi (BL #33762), UAS-mito-HA-GFP (abbreviated as UAS-mitoGFP; BL #8442), UAS-hPINK1-FLAG (BL #52004), UAS-Cdk8ORF which expresses a full-length open reading frame (F001348), UAS-Cdk8-RNAi (BL #67010), UAS-Cdk8-3xHA which expresses a full-length HA-tagged Cdk8 (F001713), UAS-Cdk8KD was obtained from Dr. J-Y Ji from Tulane University School of Medicine, UAS-CycC-RNAi (BL #33753), UAS-CycC-3xHA which expresses a full-length HA-tagged CycC (F003051), UAS-luciferase RNAi (BL #35788), UAS-CDK19WT, UAS-CDK19Y32H and UAS-CDK19T196A were generated in the previous study (Chung et al. 9). Strains with BL stock number were obtained from Bloomington Drosophila Stock Center (Bloomington, IN, USA). Strains labeled with an F initial were obtained from FlyORF Drosophila Stock Center (ZurichORFeome Project, Zurich, Switzerland).
Documentation of adult Drosophila phenotypes
Adult flies were anesthetized with CO2 gas and photographed on a Leica MZ6 dissecting stereo microscope with an attached Leica IC90 E digital camera.
Generation of UAS-CDK19ΔNLS transgenic flies
UAS-CDK19ΔNLS transgenic flies were generated as previously described78. Site-directed mutagenesis was performed with the Q5 site-directed mutagenesis kit (NEB), followed by Sanger verification. The following forward and reverse primers were used to make CDK19ΔNLS or CDK19ΔNLS −3xHA
FW- CGGCTAGGGCCTTCAGGC
RV- GTTTGGAGGCACCTGGTTCAG
Using Gateway cloning (Thermo Fisher Scientific), the CDK19 cDNA entry clone (GenBank: NM_015076.3) in the pDONR221 vector was shuttled to the pGW-attB-HA79. All UAS-cDNA Constructs were inserted into the VK37 (PBac{y[+]-attP}VK00037) docking site by ϕC31 mediated transgenesis80.
Generation of endogenously tagged Cdk8::GFP flies
The Cdk8-superfolder-GFP line (referred to as Cdk8::GFP) was generated as described43. Briefly, a fragment with gRNAs targeting 5′UTR (GTTATCGGGAGACAGCTGATTGG) and end of coding region (TTAGAAGGCTAGAAATTAGTAGG) of cdk8 and 200 bps of homology arms (corresponding to 200 bps upstream of 5’ sgRNA cut site and 200 bps downstream 3’ sgRNA cut site) were synthesized and cloned in pUC57_Kan_gw_OK2 custom vector backbone, generating a homology donor intermediate vector by Genewiz/Azenta. Fragments for the gene coding region (between 5’ sgRNA PAM sequence up to final amino acid before the stop codon), linker-sfGFP-stop codon and Scarless-DsRed were PCR amplified with primers containing overlaps and the fragments were assembled using NEB-HiFi DNA Assembly kit (New England Biolabs #E2621) using manufacturer’s instructions, in the homology donor intermediate linearized by BsaI-HF (NEB #R3535) to generate the homology donor vector for injection. Homology donor vector (250 ng/μL) was injected into y1w*; attP40(y + ){nos-Cas9(v + )}; iso5 embryos81. The resulting G0 males and females were crossed to y w flies to screen for the presence of 3XP3-DsRed. The resulting transgenics were PCR-verified with primers that amplify tagged gene-specific amplicons.
Total ATP determination
For the total ATP determination from the fly heads with indicated genotypes, we followed the protocol from ref. 82. For each set, ten heads were taken, and the total ATP was measured using an ATP determination kit (A22066, Life Technologies) as per the manufacturer’s instructions. The ATP levels were normalized with the total protein levels.
Immunostaining and quantification of mitochondria in adult fly brain
Heads from the adult flies were dissected out in cold PBS, and the immunostaining was performed as per ref. 83. Rabbit anti-Drp1 and Mouse anti-ATP5a (Abcam, ab14748) antibodies were used in 1:200 and 1:500 dilutions, respectively. The secondary antibodies, including goat anti-rabbit Alexa488 conjugated and goat anti-mouse Alexa555 conjugated, were used in 1:200 dilution. The images were acquired using a Zeiss880 confocal microscope, and the mitochondrial morphology was analyzed using Mitochondrial Network Analysis (MiNA) plug-in software in ImageJ.
Transmission electron microscopy
Adult fly retinas were processed for TEM imaging as described84,85. Samples were processed using a Ted Pella Bio Wave microwave oven with a vacuum attachment. Adult fly heads were dissected at 25 °C in 4% paraformaldehyde, 2% glutaraldehyde, and 0.1 M sodium cacodylate (pH 7.2). Samples were subsequently fixed at 4 °C for 48 h. 1% osmium tetroxide was used for secondary fixation and subsequently dehydrated in ethanol and propylene oxide, and then embedded in Embed-812 resin (Electron Microscopy Science, Hatfield, PA). 50 nm ultra-thin sections were obtained with a Leica UC7 microtome and collected on Formvar-coated copper grids (Electron Microscopy Science, Hatfield, PA). Specimens were stained with 1% uranyl acetate and 2.5% lead citrate and imaged using a JEOL JEM 1010 transmission electron microscope with an AMT XR-16 mid-mount 16 megapixel CCD camera. For quantification of ultrastructural features, electron micrographs were examined from three different animals per treatment.
Climbing assays
Flies raised at both 25 and 29 °C were subjected to a climbing assay at 3 days, 1 week, 2 weeks, and 3 weeks after eclosion raised at both 25 and 29 °C. Adult flies were transferred into an empty polystyrene vial with a label at a height of 5 cm. Prior to the assay, flies were gently tapped down to the bottom of the vial, and their climbing in 15 s was recorded and quantified. The test was repeated six times for each vial, and three vials per genotype were tested. When using either muscle or neuronal drivers, defects were only manifested in 2-week-old flies, while ubiquitous knockdown caused climbing defects by day 3.
Lifespan assay
Adult flies were maintained on standard media at 25 °C and transferred to fresh vials every 3 days. Mortality was scored daily.
Pupal lethality assay
The numbers of pupal cases were counted 3 days after most control flies had eclosed. Counting includes the number of pupal cases in total and the number of pupal cases with dead pharate adults. The test was repeated three times.
qRT-PCR
Whole larvae from the third instar larval stage or adult thoraxes were briefly washed in PBS and temporarily stored in RNAlater Stabilization solution (Invitrogen AM7020). Total RNA was extracted using RNeasy Mini Kits (Qiagen 74101). The first strand of cDNA was synthesized using the OneScript Plus cDNA Synthesis Kit (Abm G236). qRT-PCR were performed using SensiFast SYBR Lo-ROX Kit (Bioline 94005) on StepOne Real-time PCR System (Applied Biosystems). Primers used are:
cdk8 F CATCCGGGTGTTTCTGTCG
cdk8 R CAGCCCGATGGAACTTAATGAT
cycC F AGTTTCCCTACCGCACCAATC
cycC R ACAATCAAGCAGCAATCCAGG
pink1 FAAGCGAGGCTTTCCCCTAC
pink1 R GCACTACATRGACCACCGATTT
parkin F GAAGCCTCCAAGCCTCTAAATG
parkin R ACGGACTCTTTCTTCATCGGT
opa1 F CAAGCTGCGATACATCGTCC
opa1 R GCAGTCCATCCTTCCATTCC
marf F GAGACGACCACCTTTATCAACG
marf RCCACCTTCATGTGATCCCG
drp1 FACAGCCCACTCGATGATCG
drp1 RAAGCACTTCTTGGTGTGCAG
rp49 FAGCATACAGGCCCAAGATCG
rp49 RTGTTGTCGATACCCTTGGGC
Immunofluorescence staining
Larval body wall muscles and adult thoraxes were dissected in PBS and fixed in 4% paraformaldehyde (PFA) for 15 min at room temperature. Samples were washed with PBS with 0.1% Triton X-100 (PBST). After blocking with 5% BSA in PBST for 1 h at room temperature, samples were incubated with primary antibodies overnight at 4 °C. The following primary antibodies were used: rabbit anti-Drp1 (1:500; Cell Signaling D6C7), Rabbit anti-Drp1 S616 (1:500, Cell Signaling D9A1), mouse anti-HA (1:500; Abm G036), rabbit anti-CDK19 (1:200; Sigma-Aldrich SAB4301196), mouse- anti-Lamin C (1:100, DSHB LC28.26), mouse anti-ATP5α (1:500; Abcam ab14748), and rabbit anti-HA (1:500; Abcam ab9110). After washing with PBST, samples were incubated with Cy3- and/or Alexa Fluor 647-conjugated secondary antibodies (1:500; Jackson ImmunoResearch Laboratories), DAPI (final concentration: 0.2 μg per ml; Invitrogen D1306) for 2 h at room temperature. Samples were mounted in 70% glycerol in PBS after washing. Images were taken on a Nikon Air laser-scanning confocal microscope or a Zeiss LSM880 with an Airyscan confocal microscope and processed using ImageJ.
Quantification of mitochondrial morphology
Airyscan-processed images of larval body wall muscles were subjected to binary and skeletonize processes using ImageJ prior to morphological quantification. Each processed image was randomly assigned into five sections, and each section was subjected to Mitochondrial Network Analysis (MiNA) to quantify the following three parameters: number of networks (structure with at least one junction), mean length of network branches (mean branch length), and mitochondrial footprint (area). Data collected from five sections with four biological replicates were pooled together for statistics and generating box plots. Airyscan-processed images of adult indirect flight muscles were used in ImageJ for morphological quantification. The length of mitochondria is measured, where number of mitochondria are manually counted. Data collected from two sections with five biological replicates were pooled together for statistics and generating box plots.
Quantification of muscle fiber width
Airyscan-processed images were randomly assigned into five sections, and muscle fibers within the section were measured using ImageJ. Data collected from five sections were pooled together for statistics and generating box plots.
ROS staining
ROS assays were performed according to the previously described protocol for in vivo detection of ROS86. Briefly, adult thoraxes were dissected in PBS and incubated with 30 µM DHE (Cayman 12013) for 7 min in the dark at room temperature. After rinsing with PBS briefly, samples were fixed in 8% PFA and washed with PBS, each for 5 min before mounting. Images were taken within 2 days.
For the detection of ROS in adult brains, a previously described protocol was used with minor modifications87. Day 5 fly brains were dissected in Schneider’s medium (SDM). After brief washing with SDM, the brains were incubated for 5 min with 150 μL DHE (final concentration 30 μM, Invitrogen, Cat# D11347) to detect ROS. After washing (5 min × 3 times) in SDM, brains were mounted with VECTASHIELD Antifade Mounting Medium and then observed immediately with a Leica SP8X confocal microscope. Images were obtained as Z series with the same interval (4 μm) for the whole brain. Z series images were merged by ImageJ (Image-Stacks-Z projection-SUM slices), and then the fluorescence intensity was measured.
Electroretinogram
ERG recordings were performed as described in refs. 88,89. In brief, flies were glued to a slide with Elmer’s Glue. A recording electrode filled with 100 mM NaCl was placed on the eye, and a reference electrode was placed on the fly head. During the recording, a 1 s pulse of light stimulation was given, and the ERG traces of ten flies for each genotype were recorded and analyzed with WinWCP v.5.3.3 software.
Western blotting
Cells or tissues were lysed with 1× Cell Lysis Buffer (Cell Signaling Technology), supplemented with 1× Protease Inhibitors (Roche), and 1 mM phenylmethylsulfonyl fluoride. Protein lysates with 1× SDS sample buffer were resolved on SDS/PAGE and then transferred to nitrocellulose membranes. Membranes were blocked with 5% skimmed milk or 5% bovine serum albumin. The following primary antibodies were used: rabbit anti-Drp1 (1:2500; Cell Signaling D6C7), rabbit anti-pDrp1S616 (1:2500; Cell Signaling 4494), mouse anti-Myc (1:2500; Millipore clone 4A6), rat anti-HA-peroxidase (1:5000; Sigma-Aldrich clone 3F10), mouse anti-Actin (1:5000; Abcam ab3280), rabbit anti-GFP (1:5000, Thermo Fisher A11122), and mouse anti-HSP60 (1:1000; Thermo Fisher MA3-012). HRP (Jackson ImmunoResearch) conjugated secondary antibodies were used. Images were acquired by Amersham Imager 600.
Co-immunoprecipitation
Protein extracts were incubated with indicated IgG (Abcam ab104155), HA (Abcam ab214758), or Myc-agarose beads (Abcam ab1253) overnight at 4 °C. The beads were washed with lysis buffer and boiled in 4x Laemmli buffer. The supernatants were then analyzed by Western blotting. For detecting the endogenous Cdk8::GFP, protein extracts prepped from adult thoraxes were pre-cleared with IgG (Santa Cruz, sc-2343) for 1 h at 4 °C, lysates were then incubated with either IgG (Santa Cruz, sc-2343) or anti-HA (Santa Cruz, sc-7392) overnight at 4 °C. The beads were washed with lysis buffer and boiled in 4x Laemmli buffer. The supernatants were then analyzed by Western blotting.
Subcellular fractionation
All preparations were performed on ice. Subcellular fractionation was performed as described in refs. 90,91 with slight modification. S2R+ insect cells co-transfected with Cdk8 and Drp1 plasmids were resuspended and homogenized in 0.1% Triton in 200 μL 1x PBS (pH 7.4). About 30 μL of the homogenized solution was collected as the whole lysate. The rest of the solution was centrifuged at 10,000 × g for 30 s. About 30 μL of supernatant was collected as cytoplasmic fraction. To clear out the debris in the nuclear fraction, five subsequent washings with 0.1% Triton were performed, followed by centrifugation at 10000×g for 1 min per washing. Whole lysate and cytoplasmic fractions were boiled for 5 min with 4x Laemmli, whereas the nuclear fractions were boiled for 10 min with 1x Laemmli, and sonicated with 2 pulse, 5 s each.
Kinase assays
Clean cytoplasmic fractions from S2R+ insect cells containing pAWM-Cdk8 and pAWH-Drp1 from subcellular fractionations were subjected to co-immunoprecipitation with Myc-agarose beads (Abcam ab1253) overnight at 4 °C. The beads were washed with lysis buffer and centrifugated at 3000 × g for 2 min at 4 °C. Beads were kept to perform subsequent kinase assay following protocol listed in ref. 92 with a final concentration of 0.2 mM ATP added. The reactions were paused after 30 min, centrifugated at max speed for 5 min at 4 °C, and boiled for 10 min with 4x Laemmli. The level of Cdk8, Drp1, and pDrp1 were detected using mouse anti-Myc (1:2500; Millipore clone 4A6), rabbit anti-Drp1 (1:2500; Cell Signaling D6C7), rabbit anti-pDrp1S616 (1:2500; Cell Signaling 4494) on western blots, respectively.
Plasmids and cell culture
Entry clones were created from ORF by TOPO cloning (pENTR-D-TOPO, Thermo Fisher) according to the manufacturer’s instruction. The ORFs for Cdk8 [CG10572, FMO10168 (DGRC Stock 1637668; https://dgrc.bio.indiana.edu//stock/1637668; RRID:DGRC_1637668)] and Drp1 [CG3210, FI19305 (DGRC Stock 1647302; https://dgrc.bio.indiana.edu//stock/1647302; RRID:DGRC_1647302)] were from the DGRC clone collection. All ORFs were amplified by PCR using primers bearing flanking attB1 and attB2 sequences and subjected to BP recombination reactions using the pDONR/Zeo vector (Invitrogen) to generate Entry vectors. Expression vectors were obtained through LR recombination reactions using the pAWM and pAWH vectors from the DGRC Gateway collection.
Drosophila S2R+ cells (DGRC stock 150) were maintained at 25 °C in Schneider’s medium supplemented with 10% heat-inactivated FCS (Thermo Fisher Scientific). Cells were transfected with Effectene (Qiagen) according to the manufacturer’s recommendation. Twenty-four hours after transfection, cells were lysed for subsequent biochemical analysis.
Statistical analyses
We used GraphPad Prism for statistical analysis and generation of figures. Statistical analysis was done with the default settings of the software (∗ indicates p < 0.05, ∗∗ indicates p < 0.01, ∗∗∗ indicates p < 0.001, ∗∗∗∗ indicates p < 0.0001). Violin plots were generated using BoxPlotR (Spitzer et al. 2014) to demonstrate the average distribution of climbing assays.
Web resources
cNLS Mapper: https://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data are provided with this paper.
References
Fant, C. B. & Taatjes, D. J. Regulatory functions of the mediator kinases CDK8 and CDK19. Transcription 10, 76–90 (2019).
Bourbon, H. M. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 36, 3993–4008 (2008).
Firestein, R. et al. CDK8 is a colorectal cancer oncogene that regulates β-catenin activity. Nature 455, 547–551 (2008).
Peng, Y. et al. L-3-n-butylphthalide improves cognitive impairment induced by intracerebroventricular infusion of amyloid-beta peptide in rats. Eur. J. Pharmacol. 621, 38–45 (2009).
Kapoor, A. et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature 468, 1105–1111 (2010).
Broude, E. et al. Expression of CDK8 and CDK8-interacting genes as potential biomarkers in breast cancer. Curr. Cancer Drug Targets 15, 739–749 (2015).
Calpena, E. et al. De novo missense substitutions in the gene encoding CDK8, a regulator of the mediator complex, cause a syndromic developmental disorder. Am. J. Hum. Genet. 104, 709–720 (2019).
Uehara, T. et al. Pathogenesis of CDK8-associated disorder: two patients with novel CDK8 variants and in vitro and in vivo functional analyses of the variants. Sci. Rep. 10, 17575 (2020).
Chung, H. et al. De novo variants in CDK19 are associated with a syndrome involving intellectual disability and epileptic encephalopathy. Am. J. Hum. Genet. 106, 717–725 (2020).
Zarate, Y. A. et al. CDK19-related disorder results from both loss-of-function and gain-of-function de novo missense variants. Genet. Med. 23, 1050–1057 (2021).
Frank, S. et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell 1, 515–525 (2001).
Hattori, N. et al. Point mutations (Thr240Arg and Gln311Stop) in the Parkin gene. Biochem. Biophys. Res. Commun. 249, 754–758 (1998).
Kitada, T. et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 (1998).
Lücking, C. B. et al. Homozygous deletions in parkin gene in European and North African families with autosomal recessive juvenile parkinsonism. The European Consortium on Genetic Susceptibility in Parkinson’s disease and the French Parkinson’s Disease Genetics Study Group. Lancet 352, 1355–1356 (1998).
Valente, E. M. et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 304, 1158–1160 (2004).
Clark, I. E. et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441, 1162–1166 (2006).
Park, J. et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441, 1157–1161 (2006).
Narendra, D. P. et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298 (2010).
Kane, L. A. et al. PINK1 phosphorylates ubiquitin to activate parkin E3 ubiquitin ligase activity. J. Cell Biol. 205, 143–153 (2014).
Greene, J., Whitworth, A., Kuo, I., Andrews, L. & Feany, M. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl Acad. Sci. USA 100, 4078–4083 (2003).
Yang, Y. et al. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl Acad. Sci. USA 103, 10793–10798 (2006).
Deng, H., Dodson, M. W., Huang, H. & Guo, M. The Parkinson’s disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc. Natl Acad. Sci. USA 105, 14503–14508 (2008).
Poole, A. C. et al. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc. Natl Acad. Sci. USA 105, 1638–1643 (2008).
Li, X. et al. The mediator CDK8-cyclin C complex modulates Dpp signaling in Drosophila by stimulating Mad-dependent transcription. PLoS Genet. 16, e1008832 (2020).
Soanes, K. H. & Bell, J. B. The drosophila aeroplane mutant is caused by an I-element insertion into a tissue-specific teashirt enhancer motif. Genome 44, 919–928 (2001).
Chandran, S. et al. Suppression of myopathic lamin mutations by muscle-specific activation of AMPK and modulation of downstream signaling. Hum. Mol. Genet. 28, 351–371 (2019).
Beilina, A. et al. Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc. Natl Acad. Sci. USA 102, 5703 (2005).
Pickrell, A. M. & Youle, R. J. The roles of PINK1, Parkin, and mitochondrial fidelity in parkinson’s disease. Neuron 85, 257–273 (2015).
Silvestri, L. et al. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum. Mol. Genet. 14, 3477–3492 (2005).
Fillano, J. J., Goldenthal, M. J., Rhodes, C. H. & Marín-García, J. Mitochondrial dysfunction in patients with hypotonia, epilepsy, autism, and developmental delay: HEADD syndrome. J. Child Neurol. 17, 435–439 (2002).
Liesa, M. & Shirihai, O. S. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 17, 491–506 (2013).
Picard, M., Shirihai, O. S., Gentil, B. J. & Burelle, Y. Mitochondrial morphology transitions and functions: Implications for retrograde signaling? Am. J. Physiol. Regul. Integr. Comp. Physiol. 304, R393 (2013).
Pilling, A. & Saxton, W. P{mitoGFP.AP} construct and insertions. Personal communication to FlyBase. http://flybase.org/reports/FBrf0178877.htm (2004).
Cooper, K. F., Khakhina, S., Kim, S. K. & Strich, R. Stress-induced nuclear-to-cytoplasmic translocation of cyclin C promotes mitochondrial fission in yeast. Dev. Cell 28, 161–173 (2014).
Wang, K., Yan, R., Cooper, K. F. & Strich, R. Cyclin C mediates stress-induced mitochondrial fission and apoptosis. Mol. Biol. Cell 26, 1030–1043 (2015).
Ganesan, V. et al. Cyclin C directly stimulates Drp1 GTP affinity to mediate stress-induced mitochondrial hyperfission. Mol. Biol. Cell 30, 302–311 (2019).
Youle, R. J. & van der Bliek, A. M. Mitochondrial fission, fusion, and stress. Science 337, 1062–1065 (2012).
Bukau, B. & Horwich, A. L. The Hsp70 and Hsp60 chaperone machines. Cell 92, 351–366 (1998).
Verstreken, P. et al. Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron 47, 365–378 (2005).
Vos, M., Lauwers, E. & Verstreken, P. Synaptic mitochondria in synaptic transmission and organization of vesicle pools in health and disease. Front. Synaptic Neurosci. 2, 139 (2010).
Xue, J., Scotti, E. & Stoffel, M. CDK8 regulates insulin secretion and mediates postnatal and stress-induced expression of neuropeptides in pancreatic β cells. Cell Rep. 28, 2892–2904.e7 (2019).
Yanagawa, S., Lee, J. S. & Ishimoto, A. Identification and characterization of a novel line of Drosophila Schneider S2 cells that respond to wingless signaling. J. Biol. Chem. 273, 32353–32359 (1998).
Kanca, O. et al. An expanded toolkit for Drosophila gene tagging using synthesized homology donor constructs for CRISPR-mediated homologous recombination. Elife 11, e76077 (2022).
Songyang, Z. et al. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr. Biol. 4, 973–982 (1994).
van der Bliek, A. M., Shen, Q. & Kawajiri, S. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb. Perspect. Biol. 5, a011072 (2013).
Ma, R. et al. DUSP6 SUMOylation protects cells from oxidative damage via direct regulation of Drp1 dephosphorylation. Sci. Adv. 6, eaaz0361 (2020).
Han, H. et al. PINK 1 phosphorylates Drp1 S616 to regulate mitophagy‐independent mitochondrial dynamics. EMBO Rep. 21, 1–17 (2020).
Gao, Q. et al. PINK1-mediated Drp1 S616 phosphorylation modulates synaptic development and plasticity via promoting mitochondrial fission. Signal Transduct. Target. Ther. 7, 103 (2022).
Swank, D. M. Mechanical analysis of Drosophila indirect flight and jump muscles. Methods 56, 69–77 (2012).
Fernandes, J., Bate, M. & Vijayraghavan, K. Development of the indirect flight muscles of Drosophila. Development 113, 67–77 (1991).
Pesah, Y., Pham, T., Burgess, H., Middlebrooks, B. & Verstreken, P. Drosophila parkin mutants have decreased mass and cell size and increased sensitivity to oxygen radical stress. Development 131, 2183–2194 (2004).
Shiba-Fukushima, K., Inoshita, T., Hattori, N. & Imai, Y. PINK1-mediated phosphorylation of Parkin boosts Parkin activity in Drosophila. PLoS Genet. 10, e1004391 (2014).
Zhang, T., Mishra, P., Hay, B. A., Chan, D. & Guo, M. Valosin-containing protein (VCP/p97) inhibitors relieve mitofusin-dependent mitochondrial defects due to VCP disease mutants. Elife 6, e17834 (2017).
Abramov, A. Y. et al. Bioenergetic consequences of PINK1 mutations in Parkinson disease. PLoS ONE 6, e25622 (2011).
Chen, F. et al. Context-dependent responses of Drosophila intestinal stem cells to intracellular reactive oxygen species. Redox Biol. 39, 101835 (2021).
Koehler, C. L., Perkins, G. A., Ellisman, M. H. & Jones, D. L. Pink1 and Parkin regulate Drosophila intestinal stem cell proliferation during stress and aging. J. Cell Biol. 216, 2315–2327 (2017).
Tang, C. et al. PINK1-PRKN/PARK2 pathway of mitophagy is activated to protect against renal ischemia-reperfusion injury. Autophagy 14, 880–897 (2018).
Bancerek, J. et al. CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response. Immunity 38, 250–262 (2013).
Poss, Z. C. et al. Identification of mediator kinase substrates in human cells using cortistatin A and quantitative phosphoproteomics. Cell Rep. 15, 436–450 (2016).
Wang, J. et al. MARRVEL: integration of human and model organism genetic resources to facilitate functional annotation of the human genome. Am. J. Hum. Genet. 100, 843–853 (2017).
Westerling, T., Kuuluvainen, E. & Mäkelä, T. P. Cdk8 is essential for preimplantation mouse development. Mol. Cell. Biol. 27, 6177–6182 (2007).
Esposito, G. et al. Aconitase causes iron toxicity in Drosophila pink1 mutants. PLoS Genet. 9, e1003478 (2013).
Chan, D. C. Mitochondria: dynamic organelles in disease, aging, and development. Cell 125, 1241–1252 (2006).
Hoppins, S. The regulation of mitochondrial dynamics. Curr. Opin. Cell Biol. 29, 46–52 (2014).
Song, Z., Ghochani, M., McCaffery, J. M., Frey, T. G. & Chan, D. C. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol. Biol. Cell 20, 3525–3532 (2009).
Bleazard, W. et al. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1, 298–304 (1999).
Perdiz, D., Lorin, S., Leroy-Gori, I. & Poüs, C. Stress-induced hyperacetylation of microtubule enhances mitochondrial fission and modulates the phosphorylation of Drp1 at 616Ser. Cell. Signal. 39, 32–43 (2017).
Zaja, I. et al. Cdk1, PKCδ and calcineurin-mediated Drp1 pathway contributes to mitochondrial fission-induced cardiomyocyte death. Biochem. Biophys. Res. Commun. 453, 710–721 (2014).
Kashatus, J. A. et al. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol. Cell 57, 537–551 (2015).
Xu, S. et al. CaMKII induces permeability transition through Drp1 phosphorylation during chronic β-AR stimulation. Nat. Commun. 7, 13189 (2016).
Waterham, H. R. et al. A lethal defect of mitochondrial and peroxisomal fission. N. Engl. J. Med. 356, 1736–1741 (2007).
Chao, Y.-H. et al. Missense variants in the middle domain of DNM1L in cases of infantile encephalopathy alter peroxisomes and mitochondria when assayed in Drosophila. Hum. Mol. Genet. 25, 1846–1856 (2016).
Ishihara, N. et al. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 11, 958–966 (2009).
Léopold, P. & O’Farrell, P. H. An evolutionarily conserved cyclin homolog from Drosophila rescues yeast deficient in G1 cyclins. Cell 66, 1207–1216 (1991).
Lew, D. J., Dulić, V. & Reed, S. I. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell 66, 1197–1206 (1991).
Cooper, K. F., Mallory, M. J., Smith, J. B. & Strich, R. Stress and developmental regulation of the yeast C-type cyclin Ume3p (Srb11p/Ssn8p). EMBO J. 16, 4665–4675 (1997).
Duffy, J. B. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis 34, 1–15 (2002).
Marcogliese, P. C. et al. IRF2BPL is associated with neurological phenotypes. Am. J. Hum. Genet. 103, 456 (2018).
Bischof, J. et al. A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development 140, 2434–2442 (2013).
Venken, K. J. T., He, Y., Hoskins, R. A. & Bellen, H. J. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314, 1747–1751 (2006).
Kondo, S. & Ueda, R. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195, 715–721 (2013).
Yoon, W. H. et al. Loss of nardilysin, a mitochondrial co-chaperone for α-ketoglutarate dehydrogenase, promotes mTORC1 activation and neurodegeneration. Neuron 93, 115–131 (2017).
Marcogliese, P. C. et al. Loss of IRF2BPL impairs neuronal maintenance through excess Wnt signaling. Sci. Adv. 8, eabl5613 (2022).
Chung, H.-L. et al. Loss- or gain-of-function mutations in ACOX1 cause axonal loss via different mechanisms. Neuron 106, 589–606.e6 (2020).
Chung, H.-L. et al. Very-long-chain fatty acids induce glial-derived sphingosine-1-phosphate synthesis, secretion, and neuroinflammation. Cell Metab. 35, 855–874.e5 (2023).
Owusu-Ansah, E., Yavari, A. & Banerjee, U. A protocol for in vivo detection of reactive oxygen species. Protoc. Exch. 10.103, nprot.2008.23 (2008).
Park, Y.-J. et al. Phosphatidylserine synthase plays an essential role in glia and affects development, as well as the maintenance of neuronal function. iScience 24, 102899 (2021).
Ansar, M. et al. Visual impairment and progressive phthisis bulbi caused by recessive pathogenic variant in MARK3. Hum. Mol. Genet. 27, 2703–2711 (2018).
Ansar, M. et al. Bi-allelic variants in IQSEC1 cause intellectual disability, developmental delay, and short stature. Am. J. Hum. Genet. 105, 907–920 (2019).
Nabbi, A. & Riabowol, K. Rapid isolation of nuclei from cells in vitro. Cold Spring Harb. Protoc. 2015, 769–772 (2015).
Suzuki, K., Bose, P., Leong-Quong, R. Y., Fujita, D. J. & Riabowol, K. REAP: a two minute cell fractionation method. BMC Res. Notes 3, 1–6 (2010).
Swarup, S., Pradhan-Sundd, T. & Verheyen, E. M. Genome-wide identification of phospho-regulators of Wnt signaling in Drosophila. Development 142, 1502–1515 (2015).
Acknowledgements
We thank Dr. Jun-Yuan Ji for generously sharing fly reagents. We thank Hongling Pan for creating transgenic flies and Dinghui Yu for assistance and maintenance of microscopes. Drosophila stocks were obtained from the Bloomington Stock Center (NIH P40OD018537) at Indiana University and FlyORF (ZurichORFeome Project). cDNA clones were obtained from the Drosophila Genomics Resource Center (NIH 2P40OD010949). H.J.B. is supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director (U54NS093793), and by ORIP (R24OD022005 and R24OD031447). Confocal microscopy was performed in the Neurovisualization core of the BCM IDDRC (NICHD U54HD083092). H.-l.C. was supported by the Warren Alpert Foundation. J.Z.L. is supported by a Graduate Research Award from Parkinson Canada. E.M.V. is supported by funding from the Canadian Institutes for Health Research (CIHR) Project Grants (PJT 185866 and PJT 189990) and a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (RGPIN/2014-05479).
Author information
Authors and Affiliations
Contributions
J.Z.L., H.-l.C., H.J.B., and E.M.V. conceived and designed the project. J.Z.L. and C.S. performed all experiments related to the function of Cdk8 in muscle and a subset of the climbing assays with neuronal drivers, and J.Z.L. and E.M.V. analyzed the data. H.-l.C. performed most experiments related to the neuronal function of Cdk8, and H.-l.C. and H.J.B. analyzed the data. K.K.L.W. was involved in the initial investigation of Cdk8 in muscle, and K.S. carried out Drosophila protein experiments. H.-l.C. and Z.Z. processed the samples for fly TEM experiments and analyzed TEM data. D.D. performed mitochondrial staining and ATP levels in the flies, and H.-l.C. and D.D. analyzed the data. H.-l.C. and Y.-J.P. designed and analyzed DHE staining in neurons. P.C.M. and H.-l.C. generated UAS-CDK19ΔNLS transgenic flies. O.K. and C.G.B. generated the Cdk8:GFP flies, and Z.N. performed Western blots. J.Z.L., H.-l.C., H.J.B., and E.M.V. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liao, J.Z., Chung, Hl., Shih, C. et al. Cdk8/CDK19 promotes mitochondrial fission through Drp1 phosphorylation and can phenotypically suppress pink1 deficiency in Drosophila. Nat Commun 15, 3326 (2024). https://doi.org/10.1038/s41467-024-47623-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-47623-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.