Abstract
Lateral roots (LRs) dominate the overall root surface of adult plants and are crucial for soil exploration and nutrient acquisition. When grown under mild nitrogen (N) deficiency, flowering plants develop longer LRs to enhance nutrient acquisition. This response is partly mediated by brassinosteroids (BR) and yet unknown mechanisms. Here, we show that local auxin biosynthesis modulates LR elongation while allelic coding variants of YUCCA8 determine the extent of elongation under N deficiency. By up-regulating the expression of YUCCA8/3/5/7 and of Tryptophan Aminotransferase of Arabidopsis 1 (TAA1) under mild N deficiency auxin accumulation increases in LR tips. We further demonstrate that N-dependent auxin biosynthesis in LRs acts epistatic to and downstream of a canonical BR signaling cascade. The uncovered BR-auxin hormonal module and its allelic variants emphasize the importance of fine-tuning hormonal crosstalk to boost adaptive root responses to N availability and offer a path to improve soil exploration by expanded root systems in plants.
Similar content being viewed by others
Introduction
The root system of dicots is formed by one embryonically formed primary root and post-embryonically developed lateral roots (LRs) of different orders. The formation of LRs determines the horizontal expansion of a root system and the soil volume that can be exploited for nutrients and water. LR development commences with the specification of a group of xylem-pole pericycle cells in the basal meristem and continues with a series of tightly coordinated cell divisions to give rise to a dome-shaped LR primordium1,2. These steps are followed by the formation of a radially symmetrical LR meristem, which eventually penetrates the outer cell layers of the parental root and emerges to form a mature LR1,2. The development of LRs is highly plastic, responding with altered number, angle, and length to external nutrient availability and overall plant demand for nutrients3,4,5,6.
Previous studies have revealed that N availability interferes with almost every checkpoint of LR development through recruitment of mobile peptides or by activating auxin signaling and other hormonal crosstalks7,8,9,10,11,12,13. If N in the form of nitrate is accessible only to a part of the root system, LRs elongate into the nitrate-containing patch under control of the auxin-regulated transcription factor ARABIDOPSIS NITRATE REGULATED 1 (ANR1)14,15. In contrast, local supply of ammonium triggers LR emergence by enhancing radial diffusion of auxin in a pH-dependent manner16,17. These developmental processes cease when plants are exposed to severe N limitation, which forces roots to adopt a survival strategy by suppressing LR development11,18. Suppression of LR outgrowth by extremely low N availability involves NRT1.1/NPF6.3-mediated auxin transport and the CLE-CLAVATA1 peptide-receptor signaling module11,12,19. Furthermore, LR growth under N-free conditions is controlled by the MADS-box transcription factor AGL2120. Notably, external N levels that provoke only mild N deficiency, common in natural environments or low-input farming systems, induce a systemic N foraging response characterized by enhanced elongation of roots of all orders18,21,22,23. Recently, we discovered that brassinosteroid (BR) biosynthesis and signaling are required for N-dependent root elongation24,25. Although the elongation of both the primary root (PR) and LRs are induced by mild N deficiency, LRs respond differentially to BR signaling. While PR and LR responses to low N were in overall similarly attenuated in BR-deficient mutants of Arabidopsis thaliana, loss of BRASSINOSTEROID SIGNALING KINASE 3 (BSK3) completely suppressed the response of PR but not of LRs24. These results indicate that additional signaling or regulatory components mediate N-dependent LR elongation.
Using natural variation and genome-wide association (GWA) mapping, we identified genetic variation in YUC8, involved in auxin biosynthesis, as determinant for the root foraging response to low N. We show that low N transcriptionally upregulates YUC8, together with its homologous genes and with TAA1, encoding a tryptophan amino transferase catalyzing the preceding step to enhance local auxin biosynthesis in roots. Genetic analysis and pharmacological approaches allowed placing local auxin production in LRs downstream of BR signaling. Our results reveal the importance of hormonal crosstalk in LRs where BRs and auxin act synergistically to stimulate cell elongation in response to low N availability.
Results
GWAS uncovers YUC8 as determinant for LR response to low N
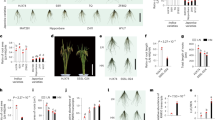
In order to identify further genetic components involved with the response of LRs to low N, we assessed LR length in a geographically and genetic diverse panel24 of 200 A. thaliana accessions grown under high N (HN; 11.4 mM N) or low N (LN; 0.55 mM N). After transferring 7-day-old seedlings precultured on sufficient N to HN or LN for 9 days, we observed substantial phenotypic variation for average LR length among tested accessions, ranging from 0.20 to 0.80 cm at HN and from 0.43 to 1.48 cm at LN (Fig. 1a, b and Supplementary Data 1). Although LR length of all examined accessions increased when plants were grown on LN (Fig. 1b), the extent of this response (i.e., the LN-to-HN ratio of average LR length) differed substantially from 22% increase as in accession Co to 188% increase in Par-3 (Fig. 1b, c). We then performed a GWA study and detected two SNPs on chromosome 4 at positions 2724898 and 14192732, respectively, that were significantly associated (false discovery rate at q = 0.05) with LR response to LN (Fig. 1d). We focused on the SNP_Chr4_14192732, as the corresponding peak was supported by adjacent markers and T-DNA insertion lines were available for all genes falling within a 20-kb supporting interval. The T-variant of this lead SNP was present in 75% of the phenotyped accessions and was associated with longer LRs under LN as compared with the A-variant (Supplementary Fig. 1a), indicating that this locus might control LR growth under LN. The SNP_Chr4_14192732 was directly located in At4g28720 (Fig. 1e), which encodes the auxin biosynthesis protein YUCCA8 (YUC8). We then analyzed T-DNA insertion lines of YUC8 and another two genes (At4g28730 and At4g28740) located within the 20-kb interval centered around the identified SNP (Fig. 1e). Knockout lines of At4g28730 and At4g28740 exhibited LN-induced LR length comparable to wild-type plants, and the expression of these two genes did not respond to LN (Supplementary Fig. 1b–e), excluding an eventual role of At4g28730 and At4g28740 in regulating LR elongation induced by mild N deficiency. By contrast, loss of YUC8 expression significantly impaired the LR response to LN (Fig. 1f, h). In two independent YUC8 mutants, average LR length was similar to wild type at HN, while at LN LRs were 25% and 18% shorter in yuc8-1 and yuc8-2 plants respectively, compared to wild-type plants. Since no significant change of PR length and LR number was observed at either N condition (Fig. 1g and Supplementary Fig. 2a), the overall decrease in total root length of yuc8 mutant plants at LN was exclusively due to decreased LR length (Supplementary Fig. 2b). Together, these results indicate that YUC8 likely underlies the trait association with SNP_Chr4_14192732.
a Representative A- and T-allele accessions of A. thaliana that show weak (Co, Ty-0, Edi-0), intermediate (Col-0), and strong (Par-3, Uod-1, Ven-1) LR elongation response to low N availability. HN, high N (11.4 mM N); LN, low N (0.55 mM N). b Reaction norms and phenotypic variation of average LR length of 200 natural accessions of A. thaliana under different N supplies. Purple diamonds represent the means of lateral root lengths for 200 accessions under each N treatment. c Frequency distribution of LR response to N availability (i.e., the ratio between LN and HN) for 200 natural accessions. d Manhattan plot for SNP associations with LR response to low N performed with vGWAS package. Negative log10-transformed P values from a genome-wide scan were plotted against positions on each of the five chromosomes of A. thaliana. Chromosomes are depicted in different colors (I to V, from left to right). The red dashed line corresponds to the Benjamini and Hochberg false-discovery rate level of q < 0.05 adjusted for multiple testing. e The 20-kb-long genomic region concentered on the lead GWA peak for LR response to low N, and genes located within this region. f–h Appearance of plants (f), primary root length (g), and average LR length (h) of wild-type (Col-0) and two yuc8 mutants. Bars represent means ± SEM. Number of individual roots analyzed in HN/LN: n = 20/19 (Col-0), 15/17 (yuc8-1), 20/20 (yuc8-2). i–k Appearance of plants (i), primary root length (j), and average LR length (k) of wild-type (Col-0) and yucQ mutant after 9 days on HN or LN. Bars represent means ± SEM. Number of individual roots analyzed in HN/LN: n = 20/21 (Col-0) and 22/17 (yucQ). Different letters in (g, h) and (j, k) indicate significant differences at P < 0.05 according to one-way ANOVA and post hoc Tukey test. Scale bars, 1 cm.
TAA1- and YUC5/7/8-dependent auxin synthesis increase LR elongation
The flavin-containing monooxygenase-like proteins of the YUCCA family have been shown to catalyze the rate-limiting step of auxin biosynthesis by converting indole-3-pyruvic acid (IPyA), produced by TAA1/TARs (Tryptophan Aminotransferase of Arabidopsis 1/ Tryptophan Aminotransferase Related proteins), into indole-3-acetic acid (IAA)26,27,28. Since YUC8 acts redundantly with its closest homologs29, we assessed root architectural traits in single mutants for two additional root-expressed YUC genes (i.e., YUC 5 and 7) and in the yuc3,5,7,8,9 quintuple mutant (yucQ). The length of PRs and LRs under N deficiency was also significantly decreased in yuc5 and yuc7 mutants (Supplementary Figs. 3 and 4). In yucQ plants, low N-induced PR and LR elongation was even completely abolished (Fig. 1i–k). Aside from defective root elongation, yucQ plants also formed significantly less LRs irrespective of the N condition (Supplementary Fig. 5). Microscopic analyses revealed that loss of the LR response to LN in yucQ plants was primarily associated with attenuated cell elongation (Fig. 2a–d). To further ascertain that auxin deficiency caused the inability of yucQ roots to respond to low N, we exogenously supplied IAA to the growth medium. Consistent with the previous studies30, PR length gradually decreased with increasing IAA supplementation in wild-type and yucQ plants (Supplementary Fig. 6a, b). However, most notably, the response of PR and especially LRs of yucQ plants to LN was fully recovered by supplying 50 nM IAA (Supplementary Fig. 6b–c). Conversely, when YUCCA-dependent auxin biosynthesis in roots of wild-type plants was suppressed with 4-phenoxyphenylboronic acid (PPBo), a potent inhibitor of YUCCA activity31, low N-induced elongation of both PR and LRs was strongly reduced (Supplementary Fig. 7).
a–b Representative confocal images of root meristems (a) and mature cells (b) of Col-0 and yucQ LRs grown under high N (HN, 11.4 mM N) or low N (LN, 0.55 mM N). Red arrowheads indicate the position of the quiescent center (QC) and the boundaries between the meristematic and elongation zones (a) or between two consecutive mature cortical cells (b). Scale bars, 50 μm. c–d Length of the meristem (c) and cortical cells (d) of LRs from Col-0 and yucQ plants grown under HN or LN. Bars represent means ± SEM. Number of individual roots or cells analyzed in HN/LN: n = 10/8 (Col-0) and 10/9 (yucQ) in (c); 34/16 (Col-0) and 45/43 (yucQ) in (d). Different letters indicate significant differences at P < 0.05 according to one-way ANOVA and post hoc Tukey test. e–h Transcript levels of YUC8 (e), YUC3 (f), YUC5 (g), and YUC7 (h) in response to HN and LN. Root samples for qPCR analysis were taken 9 days after transfer. Expression levels were assessed in whole roots by qPCR analysis and normalized to ACT2 and UBQ10. Bars represent means ± SEM (n = 4 independent biological replicates). P values relate to differences between two N conditions according to Welch’s t-test. i proYUC8-dependent GUS activity in the tips of primary root (left panel) and LR (right panel) at 9 days after transfer to HN or LN. Scale bars, 100 µm. j Representative images of mDII-ntdTomato and DII-n3xVenus in tips of mature LRs grown HN or LN and supplemented with 5 µM YUCCA activity inhibitor 4-phenoxyphenyl boronic acid (PPBo). k DII-n3xVenus/mDII-ntdTomato intensity ratio in epidermal cells of mature LRs. The experiments in (a, b) and (i, j) were repeated twice with similar results. Dots represent means ± SEM (n = 30, 25, 15, and 15 roots for HN, LN, HN-PPBo and LN-PPBo, respectively). Scale bars, 100 µm.
As the expression of TAA1 is upregulated by moderate N limitation in roots21 (Supplementary Fig. 8), we then investigated if also TAA1 is required for root growth responses to mild N deficiency. Similar to yucQ plants, low N-induced elongation of PR and LRs were also strongly impaired in two independent taa1 mutants (Supplementary Fig. 9). To further test the role of local auxin biosynthesis in roots for N-dependent root foraging responses, we supplied the polar auxin transport inhibitor N-1-naphthylphthalamic acid (NPA) to the shoots in a split-agar setup (Supplementary Fig. 10). Our results showed that LR response to low N was not significantly inhibited when shoot-to-root auxin translocation was blocked. Collectively, these results indicate that TAA1- and YUC5/7/8-mediated local auxin production in roots modulates root elongation under mild N deficiency.
Previously, it has been shown that the transcription factor AGL21 is required for sustaining LR elongation in N-free media, and that auxin accumulation in LRs and the expression of multiple YUC genes can be altered by AGL21 mutation or overexpression under non-stressed conditions20. We then investigated whether AGL21 and its close homologous gene ANR1 also control systemic stimulation of LR elongation by mild N deficiency. We found that the agl21 anr1 double mutant exhibits comparable root foraging responses to mild N deficiency as wild-type plants (Supplementary Fig. 11). These results suggest that distinct mechanisms modulate foraging versus survival responses in roots. In support of this notion, roots of yuc8 or yucQ mutants responded to N starvation similarly to wild-type plants (Supplementary Figs. 12 and 13), indicating that survival responses to low N are likely independent of YUCCA-dependent local auxin biosynthesis in roots.
Low N enhances YUC3/5/7/8 to increase auxin in LR tips
We next investigated whether external N availability regulates the expression of root-expressed YUC genes. Similar to TAA1, mRNA levels of YUC8, YUC3, YUC5 and YUC7 were also significantly upregulated by low N (Fig. 2e–h). N-dependent regulation of YUC8 was confirmed by assessing YUC8 promoter activity in the meristems of PR and LRs (Fig. 2i and Supplementary Fig. 14a, b). Whereas previous studies have shown that low N availability increases auxin levels in roots32,33,34, our results indicated that this relies on a YUCCA-dependent increase in local auxin biosynthesis. To further test this assumption, we monitored auxin accumulation with the ratiometric auxin sensor R2D235. We found that DII-n3xVenus/mDI-ntdTomato ratio decreased in both PR and LR tips of low N-grown plants, which is indicative of higher auxin accumulation (Fig. 2j, k, and Supplementary Fig. 14c, d). Inhibition of YUCCAs by the supply of PPBo to roots substantially reverted low N-induced auxin accumulation (Fig. 2j, k and Supplementary Fig. 14c, d), thus corroborating the critical role of YUCCAs in enhancing local auxin biosynthesis and stimulating root elongation under mild N deficiency.
Allelic coding variants of YUC8 determine LR foraging
Our GWA mapping and genetic analyses indicated that allelic variation in YUC8 is linked to phenotypic variation of LR growth. Expression levels of YUC8 at HN and LN or expression changes in representative natural accessions with contrasting LR responses to LN were neither significantly correlated with average LR length nor with the LR response to LN (Supplementary Fig. 15). These results suggested that YUC8-dependent natural variation under LN is likely not due to variations at the transcript level. We then searched for SNPs within YUC8’s coding sequence from 139 re-sequenced lines from our original panel and detected 17 SNPs (MAF > 5%), all of which result in synonymous substitutions, except for two SNPs (T41C and A42T) that together result in a non-synonymous substitution from leucine (L) to serine (S) at position 14 (Supplementary Data 2). This non-synonymous substitution is 14 amino acids away from the FAD-binding motif, which is critical for YUC8 activity36,37. A generalized linear model association analysis of average LR length with these polymorphic sites showed that 6 of them were significantly associated with average LR length only at LN but not at HN (Fig. 3a). These 6 SNPs allowed us to group accessions into two major haplotypes (Supplementary Data 3), with YUC8-hap A (TAGCAA) associated with longer and YUC8-hap B (CTATGG) with shorter LRs at LN (Fig. 3b). Consequently, total LR length and total root length were on average longer in YUC8-hap A than YUC8-hap B accessions (Supplementary Fig. 16).
a Association of 17 polymorphic sites (MAF > 0.05) in the coding region of single-exon gene YUC8 in 139 re-sequenced accessions with average LR length under high N (HN, 11.4 mM N; red) or low N (LN, 0.55 mM N; cyan). The x-axis shows the nucleotide position of each variant. The y-axis shows the −log10 (P-value) for the association test using a generalized linear model (GLM), with a significance level at α = 0.05 indicated with a dashed red line. The six polymorphisms selected for further analysis were projected onto a schematic representation of a YUC8 gene structure represented by a light blue arrow. b Average LR length of natural accessions representing two major YUC8 haplotypes (n = 126 and 10 accessions for haplotype A and haplotype B, respectively). Dots represent means ± SEM and P values relate to differences between two haplogroups under respective N conditions according to Welch’s t-test. c Schematic of YUC8 constructs to complement the yucQ mutant. d–g Root phenotype of transgenic allelic complementation lines at low N. Appearance of plants (d), PR length (e), average LR length (f), and total root length (g) at LN of wild-type (Col-0), yucQ and independent transgenic plants expressing sequences coding for either YUC8-haplotype A or YUC8-haplotype B under control of the YUC8Col-0 promoter. Six independent T2 lines for each construct were assessed. Two representative lines are shown for each construct. Root system architecture was assessed after 9 days. Horizontal lines show medians; box limits indicate the 25th and 75th percentiles; whiskers extend to 1.5 times the interquartile range from the 25th and 75th percentiles. Numbers below each box indicate the number of plants assessed for each genotype under the respective N condition. Different letters in (e–g) indicate significant differences at P < 0.01 according to one-way ANOVA and post hoc Tukey test. P values relate to differences between two complementing groups according to Welch’s t-test. Scale bar, 1 cm.
To test the causality of the two identified YUC8 variants, we placed the coding sequence of YUC8 from Col-0 (YUC8-hap A) or Co (YUC8-hap B) downstream of the YUC8Col-0 promoter and expressed the constructs in the yucQ mutant (Fig. 3c). We initially observed that the short PR length and decreased growth rate of yucQ plants were rescued more efficiently by expressing the YUC8-hap A variant than YUC8-hap B (Supplementary Fig. 17). We then tested whether allelic variation in YUC8 is indeed relevant for root growth in the context of N deficiency. Consistent with our haplotype analysis (Fig. 3b), T2 yucQ plants expressing YUC8-hap A displayed longer PR and LRs than those expressing YUC8-hap B (Fig. 3d–g). To rule out possible effects of differential YUC8 expression due to random genomic integration of the expression cassette, we further assessed three independent T3 homozygous lines for each variant showing comparable YUC8 expression levels (Supplementary Fig. 18a). Also in these lines complementation of PR, LR, and total root length at LN was more efficient with YUC8-hap A than with YUC8-hap B (Fig. 4a–c and Supplementary Fig. 18b). Consequently, root foraging responses induced by mild N deficiency were significantly stronger in lines expressing the YUC8-hap A variant than in those expressing YUC8-hap B (Supplementary Fig. 18c–e). Microscopic analyses suggested that the stronger LR foraging response conferred by YUC8-hap A was primarily due to increased cell elongation (Fig. 4d, e), while meristem size made a minor contribution (Fig. 4f and Supplementary Fig. 19). We then tested if the differential auxin biosynthesis drives the divergent root foraging responses between YUC8-hap A and -hap B accessions by inhibiting the activities of YUCCAs in roots with PPBo. Whereas the selected accessions exhibited the expected differential root responsiveness to low N (i.e. LN-to-HN ratio) under mock conditions, exogenous supply of PPBo to roots completely eliminated the strong foraging response of YUC8-hap A accessions (Supplementary Fig. 20). Altogether, these data corroborated that natural variation in the coding sequence of YUC8 and YUCCA-dependent root auxin accumulation determines the extent of the root foraging response to mild N deficiency.
a–c Primary root length (a), average LR length (b), and total root length (c) of wild-type (Col-0), yucQ and three independent transgenic lines expressing sequences coding for either the YUC8-hap A or YUC8-hap B under control of the YUC8Col-0 promoter. d Representative confocal images of cortical cells of mature LRs of wild-type (Col-0), yucQ and transgenic lines complemented with either YUC8 variants under control of the YUC8Col-0 promoter grown under high N (HN, 11.4 mM N) or low N (LN, 0.55 mM N). Red arrowheads indicate the boundary between two consecutive cortical cells. One representative line was shown for each construct. Scale bars, 50 μm. e–f Length of cortical cells (e) and meristems (f) of LRs of wild-type (Col-0), yucQ and complemented yucQ lines grown under HN or LN for 9 days. The experiment was repeated twice with similar results. Horizontal lines show medians; box limits indicate the 25th and 75th percentiles; whiskers extend to 1.5 times the interquartile range from the 25th and 75th percentiles. Numbers below each box indicate the number of plants assessed for each genotype under respective N condition. Different lowercase letters at HN and uppercase letters at LN indicate significant differences at P < 0.05 according to one-way ANOVA and post hoc Tukey test.
Auxin tunes LR foraging downstream of BR signaling
Our previous work showed that BR biosynthesis and signaling are involved in regulating root elongation under low N24,25. We then explored a potential interdependence and hierarchy in auxin- and BR-dependent coordination of LR elongation in response to LN. Therefore, we generated a bsk3 yuc8 double mutant, which showed significantly shorter LRs than the wild type under LN but no additive effect compared to the single mutants bsk3 and yuc8 (Fig. 5a–c). This result suggested that BSK3 and YUC8 act in the same signaling route to modulate LR elongation at LN. Consistent with our previous observation that BR sensitivity increases in N-deficient roots24, exogenous application of brassinolide (the most bioactive BR) gradually suppressed the LR response to LN of wild-type plants (Supplementary Fig. 21). However, in the yucQ mutant, the response of LRs to LN was largely insensitive to exogenous BR supplies. In contrast, the LR foraging response to LN of the BR signaling mutants bsk3 and bsk3,4,7,8 as well as of the BR biosynthesis mutant dwf4-44 was restored under exogenous application of IAA (Fig. 5d, e and Supplementary Fig. 22). These results reveal a dependency of local auxin biosynthesis in LRs on BR function and place local auxin biosynthesis downstream of BR signaling.
a–c Appearance of plants (a), primary root length (b) and average lateral root length (c) of wild-type (Col-0), bsk3, yuc8 and bsk3 yuc8 plants grown under high N (HN, 11.4 mM N) or low N (LN, 0.55 mM N). Horizontal lines show medians; box limits indicate the 25th and 75th percentiles; whiskers extend to 1.5 times the interquartile range from the 25th and 75th percentiles. Numbers below each box indicates the number of plants assessed for each genotype under the respective N condition. d Appearance of bsk3,4,7,8 mutant plants grown at HN or LN in the presence or absence of 50 nM IAA. e The LR response of bsk3 and bsk3,4,7,8 plants to low N is rescued in presence of exogenous IAA. Dots represent means ± SEM. Number of individual roots analyzed in HN/LN: n = 19/22 (mock) and 17/17 (50 nM IAA) for Col-0; 15/15 (mock) and 17/17 (50 nM IAA) for bsk3; 17/16 (mock) and 18/18 (50 nM IAA) for bsk3,4,7,8. Average LR length was assessed 9 days after transfer. f–g Transcript levels of YUC8 in bsk3,4,7,8 (f) and BZR1 loss- (bzr1) or gain-of-function (bzr1-1D) mutants (g). Expression levels were assessed in roots by qPCR and normalized to ACT2 and UBQ10. Bars represent means ± SEM (n = 4 for Col-0, bzr1, bzr1-1D, and three independent biological replicates for bsk3,4,7,8 at both N conditions). h–i Representative images (h) and ratio of mDII-ntdTomato and DII-n3xVenus fluorescence signals (i) in mature LR tips of wild-type plants grown for 7 days on HN or LN in the presence or absence of 1 µM brassinazole, a BR biosynthesis inhibitor. j–k Representative images (j) and ratio of mDII-ntdTomato and DII-n3xVenus fluorescence signals (k) in mature LR tips of Col-0/R2D2 and bzr1-1D/R2D2. In (h–j), Scale bars, 100 µm. In (h–k), DII-n3xVenus and mDII-ntdTomato fluorescence was quantified in epidermal cells of mature LRs. Dots represent means ± SEM (n = 20 roots). Different letters in (b, c, e–g) indicate significant differences at P < 0.05 according to one-way ANOVA and post hoc Tukey test.
To further explore how BR signaling regulates auxin biosynthesis, we analyzed the N-dependent expression of YUC5, YUC7, and YUC8 in the bsk3,4,7,8, bzr1, and bzr1-1D mutants. Whereas the expression of these YUC genes was not significantly altered at HN, they were not anymore upregulated by LN in bsk3,4,7,8 and bzr1 roots (Fig. 5f, g and Supplementary Fig. 23). Likewise, LN-induced upregulation of TAA1 was also lost in the bzr1 mutant (Supplementary Fig. 8). Interestingly, in bzr1-1D mutant plants, which carry a stabilized variant of the BZR1 transcription factor38, TAA1, YUC7 and YUC8 were upregulated irrespective of the N regime (Fig. 5g and Supplementary Figs. 8 and 23d). Next, we assessed if BRs stimulate auxin accumulation in LR meristems by assessing auxin levels with the R2D2 reporter after the supply of the potent BR biosynthesis inhibitor brassinazole39 (BRZ), or in the bzr1-1D mutant with constitutively active BR signaling38. Supply of 1 µM BRZ, a concentration that can largely inhibit low N-induced LR elongation24,25, increased the DII/mDII ratio under low N (Fig. 5h, i), indicating less auxin accumulation. In contrast, the DII/mDII ratio strongly decreased in LRs of bzr1-1D irrespective of available N, suggesting that constitutive activation of BR signaling can increase auxin levels in LRs (Fig. 5j, k). Taken together, these data suggest that LN-induced LR elongation relies on BR signaling-dependent upregulation of TAA1 and YUC5/7/8 expression to increase local auxin biosynthesis.
Discussion
Root developmental plasticity is crucial for plant fitness and nutrient capture. When encountering low external N availability that induces mild N deficiency, plants from several species enlarge their root systems by stimulating the elongation of LRs18,21,22,23. Here we show that coding variation in the YUC8 gene determines the extent of LR elongation under mild N deficiency and that TAA1- and YUC5/7/8-dependent local auxin biosynthesis acts downstream of BR signaling to regulate this response (Fig. 6). Our findings not only provide insights into how auxin homeostasis itself is subject to natural variation, but uncovered a previously unknown crosstalk between BRs and auxin that coordinates morphological root responses to N deficiency.
Low external N availability that results in mild N deficiency induces the expression of the BR co-receptor BAK1 (Jia et al.24) and several genes involved in BR biosynthesis (Jia et al.25). Downstream of BR signaling, an auxin biosynthesis module composed of TAA1 and YUC8 together with its homologs YUC5 and YUC7 is induced to generate more IAA in the apical meristem of LRs (blue area in LR). Upon transport to the elongation zone (blue arrows), locally generated IAA enhances cell expansion. Allelic coding variants of YUC8 in natural accessions of A. thaliana determine the extent of the root foraging response to low N by differentially modulating cell elongation (schematic representation within dashed box).
While previous studies have shown that auxin levels increase in roots of N-deficient plants32,33,34, the source of this auxin and its contribution to low N-induced root elongation still remained unresolved. Our results show that mild N deficiency stimulates local auxin accumulation in the root apical meristem by upregulating TAA1 and a set of YUCCA genes (Fig. 6). We also raised further evidence that the signaling pathways involved with root foraging responses induced by moderate N deficiency are distinct from those required to alter root growth under N starvation, i.e. in absence of N (Fig. 1f–k and Supplementary Figs. 11–13). With the help of GWA mapping, we found that natural variants of YUC8 significantly contribute to LR elongation under mild N deficiency. YUC8 belongs to the family of flavin-containing monooxygenases (FMO), which use NADPH as electron donor and FAD as cofactor to convert IPyA to IAA37. Previously, it has been shown that a subset of YUCs, including YUC8, possesses an N-terminal signal anchor and colocalizes with the endoplasmic reticulum (ER)40. Our genetic analyses showed that expression of the YUC8-hap A coding variant conferred an overall improved root growth compared to YUC8-hap B (Figs. 3, 4 and Supplementary Figs. 17–19). In a small set of accessions, we detected two mutations (T41A42→C41T42) in the coding region of YUC8 which confer a non-synonymous substitution of leucine (L) to serine (S) at position 14. Unfortunately, a quantitative assessment of the in vitro catalytic properties of the two YUC8 proteoforms has remained technically challenging, as the production of sufficient quantities of soluble proteins has failed so far. Such difficulty is common for proteins associated with the ER membrane37,41,42. While the L to S substitution found here lies outside the critical FAD domain, it could potentially affect YUC8 activity by changing hydrophilicity or providing a putative phosphorylation site. However, so far post-translational regulation of auxin biosynthesis by phosphorylation has only been reported for TAA143 but not for YUCs.
As A. thaliana colonizes a wide range of different environments, part of the genetic variation and the resulting phenotypic variation could be associated with adaptive responses to local environments44,45. For example, it has been recently shown that natural allelic variants of the auxin transport regulator EXO70A3 are associated with rainfall patterns and determine adaptation to drought conditions46. We found that the top GWAS SNP from our study is most significantly associated with temperature seasonality and that the distribution of YUC8-hap A and -hap B variants is highly associated with temperature variability (Supplementary Fig. 24), suggesting that YUC8 allelic variants may play an adaptive role under temperature fluctuations. This possibility is supported by previous findings that YUC8-dependent auxin biosynthesis is necessary to stimulate hypocotyl and petiole elongation in response to increased air temperatures47,48. However, to what extent this putative evolutionary adaptation is related to the identified SNPs in YUC8 remains to be investigated.
Our results further demonstrate that BR levels and signaling regulate local, TAA1- and YUC5/7/8-dependent auxin production especially in LRs. Microscopic analysis indicated that mild N deficiency stimulates cell elongation in LRs, a response that can be strongly inhibited by genetically perturbing auxin synthesis in roots (Fig. 2a–d). This response resembles the effect of BR signaling that we uncovered previously24 and suggested that the coordination of root foraging response to low N relies on a genetic crosstalk between BRs and auxin. These two plant hormones regulate cell expansion in cooperative or even antagonistic ways, depending on the tissue and developmental context49,50,51,52. In particular, BR has been shown to antagonize auxin signaling in orchestrating stem cell dynamics and cell expansion in the PRs of non-stressed plants49. Surprisingly, in the context of low N availability, these two plant hormones did not act antagonistically on root cell elongation. Instead, our study uncovered a previously unknown interaction between BRs and auxin in roots that resembles their synergistic interplay to induce hypocotyl elongation in response to elevated temperatures50,51,52. Genetic analysis of the bsk3 yuc8 double mutant showed a non-additive effect on LR length compared to the single mutants bsk3 and yuc8-1 (Fig. 5a–c), indicating auxin and BR signaling act in the same pathway to regulate LR elongation under low N. Whereas the exogenous supply of BR could not induce LR elongation in the yucQ mutant under low N (Supplementary Fig. 21), exogenous supply of auxin to mutants perturbed in BR signaling or biosynthesis was able to restore their LR response to low N (Fig. 5d, e and Supplementary Fig. 22). These results collectively indicate that BR signaling regulates auxin biosynthesis at low N to promote LR elongation. Indeed, the expression levels of TAA1 and YUC5/7/8 were significantly decreased at low N in BR signaling defective mutants (Fig. 5f, g and Supplementary Figs. 8 and 23). Notably, when BR signaling was perturbed or enhanced, low N-induced auxin accumulation in the root apex was significantly compromised or increased, respectively (Fig. 5h–k). Together, these results established the dependency of BR functions on auxin biosynthesis. Although our results placed local auxin biosynthesis downstream of BR signaling (Fig. 5 and Supplementary Figs. 21–23), this signaling cascade is likely not linear and may entail a positive feedback loop, as auxin has been shown to stimulate BR biosynthesis in roots by inducing DWF4 expression53. Furthermore, our data support the view that the increased auxin produced in the apical meristem of N-deficient roots does not only counterbalance the growth-suppressive effect of elevated BR levels in the root apical meristem but also directly stimulates cell expansion in the elongation zone. Future studies may address how this local, N-responsive BR-auxin module is regulated by systemic N-demand signals and why N deficiency-induced elongation of LRs is more sensitive to auxin than the PR. Interestingly, LR elongation is stimulated in cepr1 and cepr1/2 mutants54, suggesting that systemic N signaling via the CEP-CEPRs-CEPDs cascade could be involved in the regulation of this hormonal module uncovered in the present study. In the future, it will be interesting to examine whether the BR-auxin module also plays a role in root elongation under other abiotic stresses such as phosphorus deficiency or water deficit. Under any of these constraints, employing CRISPR-mediated gene editing to turn “weak” YUC8 variants into “strong” variants could provide an opportunity to increase root elongation and subsequent water and nutrient acquisition in crops.
Methods
Plant materials and growth conditions
The Arabidopsis thaliana accession Col-0 and Col-3 were used as wild-types in this study. The T-DNA insertion lines yuc8-1 (SALK_096110C, N655757), yuc8-2 (SM_3.23299, N110939), yuc5-1 (SAIL_116_C01, N860386), yuc5-2 (SALK_088618C, N672844), yuc7-1 (SALK_059832C, N659416), yuc7-2 (SALK_034074C, N680792), dwf4-44 (SAIL_882_F07, N839744), ckrc1-1 (N66987), wei8-1 (N16407), bzr1 (SALK_208661C, N2104186) and bzr1-1D (N65987), SALK_077059C (N668516) and SAIL_1286_E04C (N867481), and the reporter line R2D2 (N2105637) were purchased from Nottingham Arabidopsis Stock Center (NASC, Nottingham, United Kingdom). The bsk3, bsk3,4,7,8, agl21 anr1, and yucQ in the Col-0 background and proYUC8-GUS lines have been described in previous studies24,55,56,57. The bsk3 yuc8 double mutant was generated by crossing the bsk3 and yuc8-1 and homozygous F3 plants were selected. Homozygotes and gene transcript levels of all lines used in the current study were confirmed by PCR and qRT-PCR using primers listed in Supplementary Data 4. The mutant lines used in the present study were described in Supplementary Data 5 and the expression levels of disrupted genes were shown in Supplementary Fig. 25.
Seeds were surface-sterilized by incubation in 70% (v/v) ethanol and 0.05% (v/v) Triton X-100 for 15 min. Seeds were sown on modified half-strength MS medium (750 µM MgSO4·7H2O, 625 µM KH2PO4, 1500 µM CaCl2·2H2O, 0.055 µM CoCl2·6H2O, 0.053 µM CuCl2·2H2O, 50 µM H3BO3, 2.5 µM KI, 50 µM MnCl2. 4H2O, 0.52 µM Na2MoO4·2H2O, 15 µM ZnCl2; 75 µM Fe-EDTA) supplemented with 11.4 mM N (1 mM NH4NO3 + 9.4 mM KNO3), 0.5% (w/v) sucrose, 1% (w/v) Difco agar (Becton Dickinson) and 2.5 mM MES (pH 5.6) and then kept in the darkness at 4 °C for two days to synchronize germination. After stratification, agar plates containing seeds were placed vertically in a growth cabinet (Percival Scientific) under a 19 °C/22 °C and 14 h/10 h night/day regime with light intensity adjusted to 120 µmol photons m−2s−1. Seven-day-old seedlings of similar size were transferred to fresh plates with identical sucrose, agar, and nutrient composition as described above but supplied with either 11.4 mM (HN) or 0.55 mM (LN) N. The position of the primary root tip at the time when the seedlings were transferred from pre-culture to the N treatments was marked by black dots or small dashes. If not indicated otherwise, plants were cultivated for 9 days on these conditions. Treatments with IAA, 4-phenoxyphenyl boronic acid (PPBo), 24-epibrassinolide (BL) or brassinazole (BRZ) were performed by transferring seven-day-old seedlings to ½ MS medium supplemented with indicated concentrations of IAA (I0901, Duchefa biochemie), PPBo (CAS Number 51067-38-0, Sigma), 24-epibrassinolide (CAS Number 78821-43-9, Sigma) or brassinazole (CAS Number 280129-83-1, Sigma) dissolved in pure ethanol or DMSO. Agar plates supplied with an identical concentration of solvent served as mock treatments.
Root phenotyping, genetic mapping, and haplotype analysis
For collecting LR lengths, we grew 200 accessions (Supplementary Data 1) on HN and LN agar plates (4 individual plants per plate) and repeated the screening three times so that a total of 12 plants were finally analyzed per accession on either N condition. Roots were scanned using an Epson Expression 10000XL scanner (Seiko Epson) with a resolution of 300 dots per inch after they were clearly separated from one another on the agar plate. Total LR length was quantified with WinRhizo Pro version 2009c (Regent Instruments) and LR number was manually counted. The average LR length was calculated by dividing total LR length by LR number. Average values calculated from 12 plants per line for 200 accessions were used as phenotypic response in GWAS. We performed GWA mapping using a vGWAS package as described by Shen et al.58 and ∼215k SNP markers59,60. The significant threshold was determined at a Benjamini and Hochberg false discovery rate level of q < 0.05 for correcting multiple testing61. For the analysis of YUC8 coding sequences, we downloaded the available coding sequences and predicted amino acid sequences of 139 genome re-sequenced accessions phenotyped in our study from the 1001 Genomes Project (http://signal.salk.edu/atg1001/3.0/gebrowser.php). Sequences of 139 accessions were aligned with ClustalW 2.1 (http://bar.utoronto.ca) to extract SNPs. Only polymorphisms with minor allele frequency (MAF) > 5% were considered. YUC8-based association analysis was performed with a generalized linear model (GLM) implemented in Tassel 2.162. Six significantly associated SNPs according to YUC8-based local association analysis (P < 0.05) were taken to define YUC8 haplotypes. Haplogroups containing at least five accessions were used for comparative analysis.
Plasmid construction and transgenic complementation
For allelic complementation, we amplified a 1982-bp-long promoter region of YUC8 from genomic DNA of accession Col-0 and the open reading frames carrying the YUC8-hap A or YUC8-hap B allele from Col-0 or Co using the primers listed in Supplementary Data 4, respectively. The amplified fragments were cloned into GreenGate entry modules (pGGA000 for promoter and pGGC000 for open reading frame) and assembled in a pGREEN-IIS-based binary vector following the instructions of Lampropoulos et al.63. Plants were transformed through the floral dip method using Agrobacterium tumefaciens strain GV3101 containing the helper plasmid pSOUP64. Positive transformants were selected on agar plates supplemented with 40 mg L−1 hygromycin.
Histological and fluorescence analyses
Tissue-specific localization of YUC8 expression was investigated by histological staining of GUS activity in transgenic plants expressing proYUC8::GUS described in Hentrich et al.55. Root samples were incubated in 20 mg ml−1 (w/v) 5-bromo-4 chloro-3-indolyl-β-D-glucuronic acid (X-gluc), 100 mM NaPO4, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6 and 0.1% (v/v) Triton X-100 at 37 °C for 60-90 min in the dark. Samples were then mounted on clearing solution (chloral hydrate: water: glycerol = 8:3:1) for 3 min and imaged using Differential Interference Contrast optics on a light microscope (Axio Imager 2, Zeiss).
For the analysis of cellular traits and expression of fluorophores in LRs, we sampled the four topmost LRs from more than 10 individual plants to minimize developmental stage-dependent variations. Roots were imaged with a laser-scanning confocal microscope (LSM 780, Carl-Zeiss). Excitation and detection of fluorophores were configured as follows: Propidium iodide was excited at 561 nm and detected at 578–718 nm; Venus was excited at 514 nm and detected at 524–540 nm; tdTomato was excited at 561 nm and detected at 566–691 nm. Signal quantifications were performed with ZEN software (Carl-Zeiss).
Quantitative real-time PCR
Root tissues were collected by excision and immediately frozen in liquid N. Total RNA was extracted using the RNeasy Plant Mini Kit (Macherey-Nagel GmbH & Co KG, Germany). qRT-PCR reactions were conducted with the CFX 384TM Real-Time System (Bio-Rad, Germany) and the Go Taq qPCR Master Mix SybrGreen I (Promega) using the primers listed in Supplementary Data 4. Relative expression was calculated according to Pfaffl65 and all genes were normalized to AtACT2 and AtUBQ10 as internal references.
Climate data and statistical analysis
A subset of climate variables gathered by Hancock et al.44 was used in this study. Raw data of climate variables (19 climate, latitude and longitude) were downloaded for 113 accessions (Supplementary Data 6) from WorldClim Project (www.Worldclim.org). Climatic variables-SNP associations in Supplementary Fig. 24a, b were extracted from Hancock et al.44 and replotted.
Root traits in several genotypes were compared by one-way ANOVA followed by post hoc Tukey test at P < 0.05. Pairwise comparisons were carried out using Welch’s t-test. All statistical analyses were performed in R (version 3.6.0)66.
Data availability
The authors declare that all data supporting the findings of this study are available within the manuscript and the Supplementary files and provided in the Source Data File. Source data are provided with this paper.
References
Peret, B. et al. Arabidopsis lateral root development: an emerging story. Trends Plant Sci. 14, 399–408 (2009).
Lavenus, J. et al. Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci. 18, 450–458 (2013).
Giehl, R. F. H., Gruber, B. D. & von Wirén, N. It’s time to make changes: modulation of root system architecture by nutrient signals. J. Exp. Bot. 65, 769–778 (2014).
Giehl, R. F. H. & von Wirén, N. Root nutrient foraging. Plant Physiol. 166, 509–517 (2014).
Jia, Z. & von Wirén, N. Signaling pathways underlying nitrogen-dependent changes in root system architecture: from model to crop species. J. Exp. Bot. 71, 4393–4404 (2020).
Giehl, R. F. H., Lima, J. E. & von Wirén, N. Localized iron supply triggers lateral root elongation in Arabidopsis by altering the AUX1-mediated auxin distribution. Plant Cell 24, 33–49 (2012).
Signora, L., De Smet, I., Foyer, C. H. & Zhang, H. ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J. 28, 655–662 (2001).
Gifford, M. L., Dean, A., Gutierrez, R. A., Coruzzi, G. M. & Birnbaum, K. D. Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl Acad. Sci. USA 105, 803–808 (2008).
Tian, Q. Y., Sun, P. & Zhang, W. H. Ethylene is involved in nitrate-dependent root growth and branching in Arabidopsis thaliana. N. Phytol. 184, 918–931 (2009).
Vidal, E. A. et al. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 107, 4477–4482 (2010).
Krouk, G. et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18, 927–937 (2010).
Araya, T. et al. CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl Acad. Sci. USA 111, 2029–2034 (2014).
Bouguyon, E. et al. Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat. Plants 1, 15015 (2015).
Zhang, H. M. & Forde, B. G. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279, 407–409 (1998).
Remans, T. et al. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl Acad. Sci. USA 103, 19206–19211 (2006).
Lima, J. E., Kojima, S., Takahashi, H. & von Wirén, N. Ammonium triggers lateral root branching in Arabidopsis in an AMMONIUM TRANSPORTER1;3-dependent manner. Plant Cell 22, 3621–3633 (2010).
Meier, M., Liu, Y., Lay-Pruitt, K. S., Takahashi, H. & von Wirén, N. Auxin-mediated root branching is determined by the form of available nitrogen. Nat. Plants 6, 1136–1145 (2020).
Gruber, B. D., Giehl, R. F. H., Friedel, S. & von Wirén, N. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 163, 161–179 (2013).
Maghiaoui, A. et al. The Arabidopsis NRT1.1 transceptor coordinately controls auxin biosynthesis and transport to regulate root branching in response to nitrate. J. Exp. Bot. 71, 4480–4494 (2020).
Yu, L. H. et al. MADS-Box transcription factor AGL21 regulates lateral root development and responds to multiple external and physiological signals. Mol. Plant 7, 1653–1669 (2014).
Ma, W. et al. Auxin biosynthetic gene TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in Arabidopsis. Plant J. 78, 70–79 (2014).
Shao, A. et al. The auxin biosynthetic TRYPTOPHAN AMINOTRANSFERASE RELATED TaTAR2.1-3A Increases grain yield of wheat. Plant Physiol. 174, 2274–2288 (2017).
Qin, L. et al. Adaption of roots to nitrogen deficiency revealed by 3-D quantification and proteomic analysis. Plant Physiol. 179, 329–347 (2018).
Jia, Z., Giehl, R. F. H., Meyer, R. C., Altmann, T. & von Wirén, N. Natural variation of BSK3 tunes brassinosteroid signaling to regulate root foraging under low nitrogen. Nat. Commun. 10, 2378 (2019).
Jia, Z., Giehl, R. F. H. & von Wirén, N. The root foraging response under low nitrogen depends on DWARF1-mediated brassinosteroid biosynthesis. Plant Physiol. 183, 998–1010 (2020).
Won, C. et al. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl Acad. Sci. USA 108, 18518–18523 (2011).
Stepanova, A. N. et al. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23, 3961–3973 (2011).
Mashiguchi, K. et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl Acad. Sci. USA 108, 18512–18517 (2011).
Chen, Q. et al. Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 55, 1072–1079 (2014).
Evans, M. L., Ishikawa, H. & Estelle, M. A. Responses of Arabidopsis roots to auxin studied with high temporal resolution-comparison of wild-type and auxin-response mutants. Planta 194, 215–222 (1994).
Kakei, Y. et al. Small-molecule auxin inhibitors that target YUCCA are powerful tools for studying auxin function. Plant J. 84, 827–837 (2015).
Caba, J. M., Centeno, M. L., Fernandez, B., Gresshoff, P. M. & Ligero, F. Inoculation and nitrate alter phytohormone levels in soybean roots: differences between a supernodulating mutant and the wild type. Planta 211, 98–104 (2000).
Walch-Liu, P. et al. Nitrogen regulation of root branching. Ann. Bot. 97, 875–881 (2006).
Kiba, T., Kudo, T., Kojima, M. & Sakakibara, H. Hormonal control of nitrogen acquisition: roles of auxin, abscisic acid, and cytokinin. J. Exp. Bot. 62, 1399–1409 (2011).
Liao, C. Y. et al. Reporters for sensitive and quantitative measurement of auxin response. Nat. Methods 12, 207–210 (2015).
Hou, X. et al. Allelic analyses of the Arabidopsis YUC1 locus reveal residues and domains essential for the functions of YUC family of flavin monooxygenases. J. Integr. Plant Biol. 53, 54–62 (2011).
Dai, X. et al. The biochemical mechanism of auxin biosynthesis by an arabidopsis YUCCA flavin-containing monooxygenase. J. Biol. Chem. 288, 1448–1457 (2013).
Wang, Z. Y. et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2, 505–513 (2002).
Asami, T. et al. Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 123, 93–99 (2000).
Kriechbaumer, V., Botchway, S. W. & Hawes, C. Localization and interactions between Arabidopsis auxin biosynthetic enzymes in the TAA/YUC-dependent pathway. J. Exp. Bot. 68, 4195–4207 (2017).
Kriechbaumer, V., Wang, P., Hawes, C. & Abell, B. M. Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J. 70, 292–302 (2012).
Chourey, P. S., Li, Q. B. & Kumar, D. Sugar-hormone cross-talk in seed development: two redundant pathways of IAA biosynthesis are regulated differentially in the invertase-deficient miniature1 (mn1) seed mutant in maize. Mol. Plant 3, 1026–1036 (2010).
Wang, Q. et al. A phosphorylation-based switch controls TAA1-mediated auxin biosynthesis in plants. Nat. Commun. 11, 679 (2020).
Hancock, A. M. et al. Adaptation to climate across the Arabidopsis thaliana Genome. Science 334, 83–86 (2011).
Fournier-Level, A. et al. A map of local adaptation in Arabidopsis thaliana. Science 334, 86–89 (2011).
Ogura, T. et al. Root system depth in Arabidopsis is shaped by EXOCYST70A3 via the dynamic modulation of auxin transport. Cell 178, 400–412 (2019).
Sun, J. Q., Qi, L. L., Li, Y. N., Chu, J. F. & Li, C. Y. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. Plos Genet. 8, e1002594 (2012).
Lee, H. J. et al. FCA mediates thermal adaptation of stem growth by attenuating auxin action in Arabidopsis. Nat. Commun. 5, 5473 (2014).
Chaiwanon, J. & Wang, Z. Y. Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr. Biol. 25, 1031–1042 (2015).
Ibanez, C. et al. Brassinosteroids dominate hormonal regulation of plant thermomorphogenesis via. BZR1. Curr. Biol. 28, 303–310 (2018).
Oh, E. et al. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife 3, e03031 (2014).
Oh, E., Zhu, J. Y. & Wang, Z. Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14, 802–809 (2012).
Chung, Y. et al. Auxin stimulates DWARF4 expression and brassinosteroid biosynthesis in Arabidopsis. Plant J. 66, 564–578 (2011).
Tabata, R. et al. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346, 343–346 (2014).
Hentrich, M. et al. The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J. 74, 626–637 (2013).
Liu, Y. et al. Involvement of a truncated MADS-box transcription factor ZmTMM1 in root nitrate foraging. J. Exp. Bot. 71, 4547–4561 (2020).
Liu, G. et al. Local transcriptional control of YUCCA regulates auxin promoted root-growth inhibition in response to aluminium stress in Arabidopsis. PLoS Genet. 12, e1006360 (2016).
Shen, X., Pettersson, M., Ronnegard, L. & Carlborg, O. Inheritance beyond plain heritability: variance-controlling genes in Arabidopsis thaliana. PLoS Genet. 8, e1002839 (2012).
Atwell, S. et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465, 627–631 (2010).
Horton, M. W. et al. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat. Genet. 44, 212–216 (2012).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B-Methodol. 57, 289–300 (1995).
Bradbury, P. J. Z., Kroon, Z., Casstevens, D. E., Ramdoss, T. M. & Buckler, Y. E. S. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635 (2007).
Lampropoulos, A. et al. GreenGate - a novel, versatile, and efficient cloning system for plant transgenesis. PLoS ONE 8, e83043 (2013).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001).
Team RC. R: a language and environment for statistical computing (R Foundation for Statistical Computing, 2013).
Acknowledgements
We acknowledge the China Scholarship Council (CSC) for financial support to Z.J. (No. 201406350062) and the Deutsche Forschungsgemeinschaft for financial support to N.v.W. (WI1728/25-1) and R.F.H.G. (HE 8362/1-1). We thank Jacqueline Fuge, Annett Bieber, Elis Fraust and Lisa Gruber (Leibniz Institute of Plant Genetics and Crop Plant Research) for excellent technical assistance. We thank Zhaojun Ding (Shandong University, China) for providing seeds of yucQ mutant and Stephan Pollmann (Centro de Biotecnología y Genómica de Plantas (CBGP), Spain) for proYUC8::GUS line. We further thank Thomas Altmann and Rhonda C. Meyer (Leibniz Institute of Plant Genetics and Crop Plant Research, Germany) for providing seeds and SNP data of Arabidopsis accessions.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
Z.J. performed most experiments. R.F.H.G. helped to prepare plasmids and transgenic plants, supervised qRT-PCR analyses, and imaged roots with confocal microscope. Z.J., R.F.H.G., and N.v.W. designed experiments, analyzed the data, and wrote the manuscript. N.v.W. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Communications thanks Santosh Satbhai and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jia, Z., Giehl, R.F.H. & von Wirén, N. Local auxin biosynthesis acts downstream of brassinosteroids to trigger root foraging for nitrogen. Nat Commun 12, 5437 (2021). https://doi.org/10.1038/s41467-021-25250-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-021-25250-x
This article is cited by
-
3,4-Dichlorophenylacetic acid acts as an auxin analog and induces beneficial effects in various crops
Communications Biology (2024)
-
Plastid-localized amino acid metabolism coordinates rice ammonium tolerance and nitrogen use efficiency
Nature Plants (2023)
-
Crosstalk between brassinosteroid signaling and variable nutrient environments
Science China Life Sciences (2023)
-
A volatile producing Bacillus subtilis strain from the rhizosphere of Haloxylon ammodendron promotes plant root development
Plant and Soil (2023)
-
The use of ecological analytical tools as an unconventional approach for untargeted metabolomics data analysis: the case of Cecropia obtusifolia and its adaptive responses to nitrate starvation
Functional & Integrative Genomics (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.