Abstract
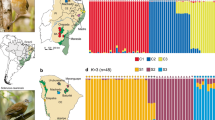
In the Anthropocene, many species are rapidly shifting their ranges in response to human-driven habitat modifications. Studying patterns and genetic signatures of range shifts helps to understand how species cope with environmental disturbances and predict future shifts in the face of global environmental change. We investigated the genetic signature of a contemporary wide-range expansion observed in the Iberian common vole Microtus arvalis asturianus shortly after a colonization event. We used mtDNA and microsatellite data to investigate patterns of genetic diversity, structure, demography, and gene flow across 57 localities covering the historical range of the species and the newly colonized area. The results showed a genetic footprint more compatible with a true range expansion (i.e. the colonization of previously unoccupied areas), than with a model of “colonization from within” (i.e. local expansions from small, unnoticed populations). Genetic diversity measures indicated that the source population was likely located at the NE of the historical range, with a declining gradient of genetic diversity towards the more recently invaded areas. At the expansion front, we observed the greatest gene flow and smallest pairwise differences between nearby localities. Both natural landscape features (rivers) and recent anthropogenic barriers (roads, railways) explained a large proportion of genetic variance among populations and had a significant impact on the colonization pathways used by voles.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Alcala N, Goudet J, Vuilleumier S (2014) On the transition of genetic differentiation from isolation to panmixia: what we can learn from GST and D. Theor Popul Biol 93:75–84
Alda F, Doadrio I (2014) Spatial genetic structure across a hybrid zone between European rabbit subspecies. PeerJ 2:e582
Angert AL, Crozier LG, Rissler LJ, Gilman SE, Tewksbury JJ, Chunco AJ (2011) Do species’ traits predict recent shifts at expanding range edges? Ecol Lett 14:677–689
Austerlitz F, Jung-Muller B, Godelle B, Gouyon P-H (1997) Evolution of coalescence times, genetic diversity and structure during colonization. Theor Popul Biol 51:148–164
Baca M, Popović D, Baca K, Lemanik A, Doan K, Horáček I et al. (2020) Diverse responses of common vole (Microtus arvalis) populations to Late Glacial and Early Holocene climate changes – Evidence from ancient DNA. Quat Sci Rev 233:106239
Baca M, Popović D, Lemanik A, Bañuls-Cardona S, Conard NJ, Cuenca-Bescós G et al. (2023) Ancient DNA reveals interstadials as a driver of common vole population dynamics during the last glacial period. J Biogeogr 50:183–196
Beerli P, Felsenstein J (2001) Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. PNAS 98:4563–4568
Bellard C, Bertelsmeier C, Leadley P, Thuiller W, Courchamp F (2012) Impacts of climate change on the future of biodiversity. Ecol Lett 15:365–377
Benítez-López A, Alkemade R, Verweij PA (2010) The impacts of roads and other infrastructure on mammal and bird populations: a meta-analysis. Biol Conserv 143:1307–1316
Bohonak AJ (1999) Dispersal, gene flow, and population structure. Q Rev Biol 74:21–45
Brodie JF (2016) Synergistic effects of climate change and agricultural land use on mammals. Front Ecol Environ 14:20–26
Caplat P, Edelaar P, Dudaniec RY, Green AJ, Okamura B, Cote J et al. (2016) Looking beyond the mountain: dispersal barriers in a changing world. Front Ecol Environ 14:261–268
Conord C, Gurevitch J, Fady B (2012) Large-scale longitudinal gradients of genetic diversity: a meta-analysis across six phyla in the Mediterranean basin. Ecol Evol 2:2600–2614
Corander J, Marttinen P (2006) Bayesian identification of admixture events using multilocus molecular markers. Mol Ecol 15:2833–2843
Crowl TA, Crist TO, Parmenter RR, Belovsky G, Lugo AE (2008) The spread of invasive species and infectious disease as drivers of ecosystem change. Front Ecol Environ 6:238–246
Dakin EE, Avise JC (2004) Microsatellite null alleles in parentage analysis. Heredity 93:504–509
D’Amico M, Ascensão F, Fabrizio M, Barrientos R, Gortázar C (2018) Twenty years of Road ecology: a topical collection looking forward for new perspectives. Eur J Wildl Res 64:26
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and high-performance computing. Nat Methods 9:772
Delibes M, Brunet-Lecomte P (1980) Presencia del Topillo Campesino ibérico. Microtus arvalis asturianus (Miller, 1908) en la Meseta del Duero. Doñana Acto Vert 7:120–123
Delibes J (1989) Plagas de topillos en España. Quercus 35:17–20
Di Rienzo A, Peterson AC, Garza JC, Valdes AM, Slatkin M, Freimer NB (1994) Mutational processes of simple-sequence repeat loci in human populations. Proc Natl Acad Sci USA 91:3166–3170
Domínguez JC, Calero-Riestra M, Olea PP, Malo JE, Burridge CP, Proft K et al. (2021) Lack of detectable genetic isolation in the cyclic rodent Microtus arvalis despite large landscape fragmentation owing to transportation infrastructures. Sci Rep. 11:12534
Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973
Duckworth RA (2008) Adaptive dispersal strategies and the dynamics of a range expansion. Am Naturalist 172:S4–S17
Dupanloup I, Schneider S, Excoffier L (2002) A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11:2571–2581
Engering A, Hogerwerf L, Slingenbergh J (2013) Pathogen–host–environment interplay and disease emergence. Emerg Microbes Infect 2:1–7
Estrada A, Morales-Castilla I, Caplat P, Early R (2016) Usefulness of Species Traits in Predicting Range Shifts. Trends Ecol Evol. 31:190–203
Excoffier L, Foll M, Petit RJ (2009) Genetic consequences of range expansions. Annu Rev Ecol Evol Syst 40:481–501
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Excoffier L, Ray N (2008) Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol Evol 23:347–351
Forsman A, Merilä J, Ebenhard T (2011) Phenotypic evolution of dispersal-enhancing traits in insular voles. Proc Biol Sci 278:225–232
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915
Fuller T, Bensch S, Müller I, Novembre J, Pérez-Tris J, Ricklefs RE et al. (2012) The ecology of emerging infectious diseases in migratory birds: an assessment of the role of climate change and priorities for future research. EcoHealth 9:80–88
García JT, Domínguez‐Villaseñor J, Alda F, Calero‐Riestra M, Olea PP, Fargallo JA et al. (2020) A complex scenario of glacial survival in Mediterranean and continental refugia of a temperate continental vole species (Microtus arvalis) in Europe. J Zool Syst Evolut Res 58:459–474
García J, Morán-Ordóñez A, García JT, Calero-Riestra M, Alda F, Sanz J et al. (2021) Current landscape attributes and landscape stability in breeding grounds explain genetic differentiation in a long-distance migratory bird. Anim Conserv 24:120–134
Gaston KJ (2009) Geographic range limits of species. Proc R Soc B: Biol Sci 276:1391–1393
Gauffre B, Estoup A, Bretagnolle V, Cosson J (2008) Spatial genetic structure of a small rodent in a heterogeneous landscape. Mol Ecol 17:4619–4629
Gauffre B, Galan M, Bretagnolle V, Cosson J (2007) Polymorphic microsatellite loci and PCR multiplexing in the common vole, Microtus arvalis. Mol Ecol Notes 7:830–832
Gerlach G, Musolf K (2000) Fragmentation of landscape as a cause for genetic subdivision in bank voles. Conserv Biol 14:1066–1074
Gilbert KJ, Sharp NP, Angert AL, Conte GL, Draghi JA, Guillaume F et al. (2017) Local adaptation interacts with expansion load during range expansion: maladaptation reduces expansion load. Am Naturalist 189:368–380
Gómez A, Lunt DH (2007) Refugia within refugia: patterns of phylogeographic concordance in the Iberian Peninsula. In: Phylogeography of southern European refugia, Springer, pp 155–188
González de Molina M, Soto Fernández D, Infante-Amate J, Aguilera E, Vila Traver J, Guzmán GI (2017) Decoupling food from land: the Evolution of Spanish agriculture from 1960 to 2010. Sustainability 9:2348
González-Esteban J, Villate I, Gosálbez J (1995) Expansión del área de distribución de Microtus arvalis asturianus Miller, 1908 (Rodentia, Arvicolidae) en la Meseta Norte (España). Doñana, Acta Vertebrata 22:106–110
Goodsman DW, Cooke B, Coltman DW, Lewis MA (2014) The genetic signature of rapid range expansions: how dispersal, growth and invasion speed impact heterozygosity and allele surfing. Theor Popul Biol 98:1–10
Goudet J (2005) hierfstat, a package for r to compute and test hierarchical F-statistics. Mol Ecol Notes 5:184–186
Hastings A, Cuddington K, Davies KF, Dugaw CJ, Elmendorf S, Freestone A et al. (2005) The spatial spread of invasions: new developments in theory and evidence. Ecol Lett 8:91–101
Heppenheimer E, Cosio DS, Brzeski KE, Caudill D, Van Why K, Chamberlain MJ et al. (2018) Demographic history influences spatial patterns of genetic diversity in recently expanded coyote (Canis latrans) populations. Heredity 120:183–195
Hewitt G (2000) The genetic legacy of the Quaternary ice ages. Nature 405:907–913
Hulme PE, Bacher S, Kenis M, Klotz S, Kühn I, Minchin D et al. (2008) Grasping at the routes of biological invasions: a framework for integrating pathways into policy. J Appl Ecol 45:403–414
Huth G, Haegeman B, Pitard E, Munoz F (2015) Long-distance rescue and slow extinction dynamics govern multiscale metapopulations. Am Naturalist 186:460–469
Ibrahim KM, Nichols RA, Hewitt GM (1996) Spatial patterns of genetic variation generated by different forms of dispersal during range expansion. Heredity 77:282–291
Ishibashi Y, Saitoh T, Abe S, Yoshida MC (1997) Cross-species amplification of microsatellite DNA in Old World microtine rodents with PCR primers for the gray-sided vole, Clethrionomys rufocanus. Mammal Study 22:5–10
Jareño D, Viñuela J, Luque-Larena JJ, Arroyo L, Arroyo B, Mougeot F (2015) Factors associated with the colonization of agricultural areas by common voles Microtus arvalis in NW Spain. Biol Invasions 17:2315–2327
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Keenan K, McGinnity P, Cross TF, Crozier WW, Prodöhl PA (2013) diveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol Evol 4:782–788
Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A (2019) RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35:4453–4455
Ledevin R, Millien V (2013) Congruent morphological and genetic differentiation as a signature of range expansion in a fragmented landscape. Ecol Evol 3:4172–4182
Legault G, Bitters ME, Hastings A, Melbourne BA (2020) Interspecific competition slows range expansion and shapes range boundaries. Proc Natl Acad Sci USA 117:26854–26860
Legendre P, Anderson MJ (1999) Distance-based redundancy analysis: testing multispecies responses in multifactorial ecological experiments. Ecol Monogr 69:1–24
Lenoir J, Svenning J-C (2015) Climate-related range shifts – a global multidimensional synthesis and new research directions. Ecography 38:15–28
Luikart G, Cornuet J-M (1998) Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conserv Biol 12:228–237
Luque-Larena JJ, Mougeot F, Vinuela J, Jareno D, Arroyo L, Lambin X et al. (2013) Recent large-scale range expansion and outbreaks of the common vole (Microtus arvalis) in NW Spain. Basic Appl Ecol 14:432–441
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Miller MP (2005) Alleles in space (AIS): computer software for the joint analysis of interindividual spatial and genetic information. J Heredity 96:722–724
Miller TEX, Angert AL, Brown CD, Lee-Yaw JA, Lewis M, Lutscher F et al. (2020) Eco-evolutionary dynamics of range expansion. Ecology 101:e03139
Mooney HA, Cleland EE (2001) The evolutionary impact of invasive species. Proc Natl Acad Sci USA 98:5446–5451
Morales C, Ortega Villazán MT (2002) Inundaciones en Castilla y león. Ería: Revista cuatrimestral de geografía, ISSN 0211-0563, No 59, 2002, pp 305–332
Mougeot F, Lambin X, Rodríguez‐Pastor R, Romairone J, Luque‐Larena J-J (2019) Numerical response of a mammalian specialist predator to multiple prey dynamics in Mediterranean farmlands. Ecology 100:e02776
Niethammer J, Winking H (1971) Die spanische Feldmaus (Microtus arvalis asturianus Miller, 1908). Bonn Zoologisches Beitrage 19:189–197
Norén K, Statham MJ, Ågren EO, Isomursu M, Flagstad Ø, Eide NE et al. (2015) Genetic footprints reveal geographic patterns of expansion in Fennoscandian red foxes. Glob Change Biol 21:3299–3312
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D et al. (2019) vegan: Community Ecology Package. R package version 2.5-6. https://CRAN.R-project.org/package=vegan
O’Regan HJ (2008) The Iberian Peninsula–corridor or cul-de-sac? Mammalian faunal change and possible routes of dispersal in the last 2 million years. Quat Sci Rev 23:2136–2144
Pacifici M, Rondinini C, Rhodes JR, Burbidge AA, Cristiano A, Watson JEM et al. (2020) Global correlates of range contractions and expansions in terrestrial mammals. Nat Commun 11:2840
Parmesan C (2006) Ecological and evolutionary responses to recent climate change. Annu Rev Ecol, Evol, Syst 37:637–669
Paulose J, Hallatschek O (2020) The impact of long-range dispersal on gene surfing. Proc Natl Acad Sci USA 117:7584–7593
Pauls SU, Nowak C, Bálint M, Pfenninger M (2013) The impact of global climate change on genetic diversity within populations and species. Mol Ecol 22:925–946
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Pecl GT, Araújo MB, Bell JD, Blanchard J, Bonebrake TC, Chen I-C et al. (2017) Biodiversity redistribution under climate change: Impacts on ecosystems and human well-being. Science 355:eaai9214
Pilkington MM, Wilder JA, Mendez FL, Cox MP, Woerner A, Angui T et al. (2008) Contrasting signatures of population growth for mitochondrial DNA and Y chromosomes among human populations in Africa. Mol Biol Evol 25:517–525
Piry S, Luikart G, Cornuet J-M (1999) Computer note. BOTTLENECK: a computer program for detecting recent reductions in the effective size using allele frequency data. J Hered 90:502–503
Rambaut A, Suchard MA, Xie D, Drummond AJ (2016) Tracer v1. 6. 2014 Available at beast. bio. ed. ac. uk/Tracer. Accessed August 1
Ramos-Onsins SE, Rozas J (2002) Statistical properties of new neutrality tests against population growth. Mol Biol Evol 19:2092–2100
Rey JM (1973) Notas sobre mastozoología ibérica. 1.- Las características biométricas y morfológicas del topillo campesino, Microtus arvalis asturianus, del Sistema Ibérico (Mammalia, Rodentia). Boletín de la Real Soc Española de Historia Nat (Biolía) 71:283–297
Rodríguez-Pastor R, Luque-Larena JJ, Lambin X, Mougeot F (2016) “Living on the edge”: The role of field margins for common vole (Microtus arvalis) populations in recently colonised Mediterranean farmland. Agriculture, Ecosyst Environ 231:206–217
Rowe KC, Rowe KMC, Tingley MW, Koo MS, Patton JL, Conroy CJ et al. (2015) Spatially heterogeneous impact of climate change on small mammals of montane California. Proc R Soc B: Biol Sci 282:20141857
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE et al. (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34:3299–3302
Rytwinski T, Fahrig L (2013) Why are some animal populations unaffected or positively affected by roads? Oecologia 173:1143–1156
Salzburger W, Ewing GB, Haeseler AV (2011) The performance of phylogenetic algorithms in estimating haplotype genealogies with migration. Mol Ecol 20:1952–1963
Sánchez-Guillén RA, Córdoba-Aguilar A, Hansson B, Ott J, Wellenreuther M (2016) Evolutionary consequences of climate-induced range shifts in insects. Biol Rev 91:1050–1064
Sexton JP, McIntyre PJ, Angert AL, Rice KJ (2009) Evolution and ecology of species range limits. Annu Rev Ecol, Evol, Syst 40:415–436
Sherpa S, Després L (2021) The evolutionary dynamics of biological invasions: A multi-approach perspective. Evolut Appl 14:1463–1484
Shine R, Brown GP, Phillips BL (2011) An evolutionary process that assembles phenotypes through space rather than through time. Proc Natl Acad Sci USA 108:5708–5711
Slatkin M, Excoffier L (2012) Serial founder effects during range expansion: a spatial analog of genetic drift. Genetics 191:171–181
Stojak J, Wójcik JM, Ruczyńska I, Searle JB, McDevitt AD (2016) Contrasting and congruent patterns of genetic structuring in two Microtus vole species using museum specimens. Mammal Res 61:141–152
Stojak J, Borowik T, Górny M, McDevitt AD, Wójcik JM (2019a) Climatic influences on the genetic structure and distribution of the common vole and field vole in Europe. Mammal Res 64:19–29
Stojak J, Tarnowska E (2019b) Polish suture zone as the goblet of truth in post-glacial history of mammals in Europe. Mammal Res 64:463–475
Suckling DM, Conlong DE, Carpenter JE, Bloem KA, Rendon P, Vreysen MJB (2017) Global range expansion of pest Lepidoptera requires socially acceptable solutions. Biol Invasions 19:1107–1119
Sundqvist L, Keenan K, Zackrisson M, Prodöhl P, Kleinhans D (2016) Directional genetic differentiation and relative migration. Ecol Evol 6:3461–3475
Svenning J-C, Gravel D, Holt RD, Schurr FM, Thuiller W, Münkemüller T et al. (2014) The influence of interspecific interactions on species range expansion rates. Ecography 37:1198–1209
Swaegers J, Mergeay J, Therry L, Larmuseau MHD, Bonte D, Stoks R (2013) Rapid range expansion increases genetic differentiation while causing limited reduction in genetic diversity in a damselfly. Heredity 111:422–429
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585
Tougard C, Renvoisé E, Petitjean A, Quéré J-P (2008) New insight into the colonization processes of common voles: inferences from molecular and fossil evidence. PLoS ONE 3:e3532
Wallingford PD, Morelli TL, Allen JM, Beaury EM, Blumenthal DM, Bradley BA et al. (2020) Adjusting the lens of invasion biology to focus on the impacts of climate-driven range shifts. Nat Clim Chang 10:398–405
Weir B, Cockerham C (1984) Weir BS, Cockerham CC. Estimating F-Statistics for the Analysis of Population-Structure. Evolution 38:1358–1370
Acknowledgements
We are very grateful to students and friends for their help during the fieldwork, with special thanks to Iván García, Xurxo Piñeiro, Fernando Arce, Julián Núñez, and Vega Santos. Thanks to lab technicians, specially to María Santoro. Samples from Campo Azálvaro were kindly provided by Juan Antonio Fargallo (MNCN, CSIC), who also provided insightful comments on the manuscript. Thanks are also due to GREFA for the support and cooperation the landowners who allowed us access to their properties, and the communities and mayors from the villages of Villalar de Los Comuneros, Boada de Campos, San Martín de Valderaduey and Fuentes de Nava. This work was supported by I + D National Plan Projects of the Spanish Ministry of Economy, Industry and Competitiveness (CGL2011-30274, CGL2015-71255-P, CGL2013-42451-P) co-funded by the European Regional Development Fund (FEDER, EU), and the Fundación BBVA Research Project TOPIGEPLA (2014 call). J Martínez-Padilla was funded by ARAID foundation and currently by Science and Education Ministry (PID2019-104835GB-100). Julio Domínguez was supported by a predoctoral grant: “Programa Talento Formación” funded by Fondo Social Europeo (FSE) and Castilla La Mancha regional government (JCCM) (ref: SBPLY/16/180501/000205). Julio Domínguez was also supported by Margarita Salas fellowship funded by NextGenerationEU, Ministry of Universities and Recovery, Transformation and ResiliencePlan, through a call from Castilla-La Mancha University. We thank three anonymous reviewers for their constructive feedback.
Author information
Authors and Affiliations
Contributions
JTG, JCD, JVM, PPO, JMP, JJO and JH conceived the study. All authors collected samples. JCD and MCR performed the lab work. JCD, MCR and JTG analysed the genetic data. JCD and JTG led the writing and the rest of authors contributed to edit and review the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Ben Evans.
Supplementary information
41437_2023_613_MOESM1_ESM.docx
Genetic footprints of a rapid and large-scale range expansion: the case of cyclic common vole in Spain. Supplemental Material
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Domínguez, J.C., Alda, F., Calero-Riestra, M. et al. Genetic footprints of a rapid and large-scale range expansion: the case of cyclic common vole in Spain. Heredity 130, 381–393 (2023). https://doi.org/10.1038/s41437-023-00613-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-023-00613-w