Abstract
Species distributed across climatic gradients will typically experience spatial variation in selection, but gene flow can prevent such selection from causing population genetic differentiation and local adaptation. Here, we studied genomic variation of 415 individuals across 34 populations of the common wall lizard (Podarcis muralis) in central Italy. This species is highly abundant throughout this region and populations belong to a single genetic lineage, yet there is extensive phenotypic variation across climatic regimes. We used redundancy analysis to, first, quantify the effect of climate and geography on population genomic variation in this region and, second, to test if climate consistently sorts specific alleles across the landscape. Climate explained 5% of the population genomic variation across the landscape, about half of which was collinear with geography. Linear models and redundancy analyses identified loci that were significantly differentiated across climatic regimes. These loci were distributed across the genome and physically associated with genes putatively involved in thermal tolerance, regulation of temperature-dependent metabolism and reproductive activity, and body colouration. Together, these findings suggest that climate can exercise sufficient selection in lizards to promote genetic differentiation across the landscape in spite of high gene flow.
Similar content being viewed by others
Introduction
Understanding how the spatial distribution of environments shapes population differentiation is a shared goal of biogeography, ecology, and evolutionary biology. The balance between selection and gene flow determines the extent to which populations will diverge genetically along environmental gradients and exhibit adaptation to local environmental conditions (Savolainen et al. 2013; Tigano and Friesen 2016; Yeaman and Otto 2011). Local adaptation is common in nature (e.g., Fraser et al. 2011; Halbritter et al. 2018; Hargreaves et al. 2020) and can occur even with high levels of gene flow if selection is strong (Yeaman and Otto 2011; Savolainen et al: 2013; Tigano and Friesen 2016). When phenotypic variation is polygenic, populations can exhibit pronounced adaptive phenotypic divergence with weak geographic genetic differentiation (Yeaman 2015). However, spatial selection mosaics in regions with high gene flow will also tend to promote a genetic architecture with a few divergent genes of large effect (Yeaman and Whitlock 2011), and these genes can therefore exhibit strong genetic differentiation across the landscape.
Climatic gradients commonly impose strong divergent selection across the landscape and can promote adaptive genetic differentiation (Keller et al. 2013; Halbritter et al. 2018). Empirical studies of plants and animals have identified population genetic differentiation across climatic regimes, even over small geographic scales, and loci that reliably associate with temperature, precipitation, and seasonality (e.g., Yeaman et al. 2016; Gibson and Moyle 2020). Adaptive divergence is facilitated when there are severe restrictions on dispersal imposed by geography (e.g., terrestrial vertebrates on islands; Bassitta et al. 2021). However, divergence can also occur in the absence of such barriers if dispersal distances are short, which is commonly the case in small vertebrates like lizards (Clobert et al. 2012; Olsson and Shine 2003; Warner and Shine 2008). As a result, even highly abundant and continuously distributed lizard species can show substantial variation in morphology and colouration across climatic regimes (Ortega et al. 2019; Ruiz Miñano et al. 2021). Furthermore, common garden experiments suggest that local physiological adaptation along climatic gradients is very common (Pettersen 2020). In line with these phenotypic effects, genotype-environment associations have been able to identify genetic differentiation of particular loci associated with climate (e.g., Rodríguez et al. 2017; Prates et al. 2018; Farleigh et al. 2021; see also Campbell-Staton et al. 2016, 2017, 2020). Yet, it remains poorly understood to what extent climate is able to cause genome-wide population differentiation when the opportunity for gene flow is high.
Common wall lizards (Podarcis muralis) in central Italy are well suited to quantify the degree of population genetic differentiation across climatic regimes in the absence of physical barriers to gene flow, and to investigate if local adaptation is accompanied by differentiation at particular loci. The climatic gradients in central Italy can be steep, with a hot, dry Mediterranean climate on the western coast and Tuscan hills, transitioning into a cooler and wetter climate in the Apennine Mountains (Fig. 1; Ruiz Miñano et al. 2021). Podarcis muralis is highly abundant throughout this region, but is absent from the hottest and driest locations and occurs only sparsely above 2000 metres elevation (Sindaco et al. 2006). There are several reasons to expect that common wall lizards are experiencing strong local selection as a result of climate. Firstly, climate commonly imposes selection on reproductive biology with lizards typically differing consistently in reproductive characteristics across latitudes and altitudes (e.g., Telemeco et al. 2010; Horvathova et al. 2013; reviewed in Uller and While 2015). Secondly, cool climate imposes selection on the thermal physiology of embryos with, for example, developmental rate evolving in response to modest climatic differences (Oufiero and Angilletta 2006; reviewed in Pettersen 2020) and very rapidly (including in P. muralis; Feiner et al. 2018; While et al. 2015b).
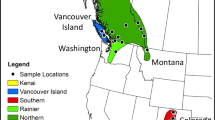
Top panel: Sampling locations in central Italy (see Table S1 for population acronyms and geographic information). Bottom panels: The spatial distribution of three climatic principal components scores, interpolated from values at the sampling locations.
In addition to the patterns of adaptive divergence in thermal physiology and life history under natural selection, common wall lizards in this region also show remarkable (genetically determined) variation in body size, shape, and colour. This variation likely results from climatic effects on the strength of sexual selection (or the balance between sexual and natural selection; MacGregor et al. 2017; While et al. 2015a; Yang et al. 2020; Ruiz Miñano et al. 2021). Highly exaggerated colours, large heads, and heavy bodies occur in hot and dry areas, whereas lizards in cooler, more seasonal climate are lighter and exhibit the brown, dull, phenotype characteristic of other genetic lineages of the same species (Ruiz Miñano et al. 2021). Yet, it is currently unknown to what extent these patterns of local phenotypic adaptation to climate are accompanied by genetic differentiation.
To quantify the extent to which climate has caused population genetic differentiation across the landscape, we used reduced representation sequencing and distance-based spatial models. We first investigated the evidence for genetic structure according to climate and geography, and quantified their unique and shared contribution to the overall genetic variation within this region. Second, we used genotype-environment association (GEA) analyses to investigate the evidence for, and distribution of, putative signatures of climatic selection in the genome.
Methods
Study system and data collection
The common wall lizard (Podarcis muralis) is a small lizard occurring throughout southern Europe (Schulte 2008). While this species has a monotypic brown phenotype across most of its range, there is a pronounced phenotypic variation across central Italy, especially in Latium, Tuscany, and Umbria (Böhme 1986; Ruiz Miñano et al. 2021). This has stimulated the description of a number of subspecies (Böhme 1986), but it is now clear that lizards throughout this area belong to a single evolutionary lineage (the ‘Tuscan’ or ‘[Central] Italian’ lineage, here referred to as the ‘IT’ lineage; Yang et al. 2018, 2022). In the northwest of the IT lineage’s distribution, there is a well-characterised hybrid zone with the distantly related ‘Southern Alps’ lineage and, further north, the river Po forms a natural barrier to gene flow with other lineages (for details, see Yang et al. 2018, 2020, 2022).
Between 2012 and 2018, we collected tail-tip tissue samples of common wall lizards from 71 locations across central and northern Italy (Fig. 1; Table S1; see below and Ruiz Miñano et al. 2021). We refer to each location as a ‘population’. The genomic data for each population was derived from up to fifteen individuals (typically 14; seven females and seven males), using double-digest restriction site-associated DNA sequencing (ddRAD-Seq) on an Illumina HiSeq 2500 platform (Novogene; Hong Kong). Library preparation for ddRAD-Seq data was implemented following the protocol in Peterson et al. (2012) and Yang et al. (2018). In brief, DNA was digested by the restriction enzymes EcoRI High-Fidelity and MspI (New England Biolabs, USA; no methylation sensitive). We used 500 ng of DNA for each individual and amplified with Q5 High-Fidelity DNA Polymerase at selected size of 300–700 bp (New England Biolabs, USA) with 12 cycles. The libraries were distributed and sequenced paired-end across a total of five runs with read length of 150 bp. We obtained approximately 1 Gb for each sample, with expected read depth of 30×. Firstly, sequence reads with low-quality score (Phred score <30), ambiguous base calls, and incomplete barcode calls were removed using “process_radtag” module in STACKS v2.41 (Catchen et al. 2013). We used STACKS to obtain single nucleotide polymorphisms (SNPs) using “ref_map” pipeline (Catchen et al. 2013) by mapping clean reads to the reference genome of P. muralis (PodMur_1.0; Andrade et al. 2019). The total assembly of the reference genome is 1.51 Gb with 2162 contig, the N50 size is 92.4 Mb, and the L50 size is 7. SNPs were estimated under a Marukilow model (Maruki and Lynch 2017) with p-value 0.05. SNPs with depth <10 were removed as low‐depth loci, and SNPs with depth >95th percentile were removed to avoid PCR duplicates, possible paralogs, and SNPs from high complexity regions (Fan et al. 2016). The individuals with average depth of coverage of <10 for all SNPs were also removed from the data set. PLINK (Chang et al. 2015) was used to filter the SNPs with minimum minor allele frequency (MAF) < 0.05, and with missing rate > 0.1 in each population. Finally, we excluded individuals with genotyping rate < 70% across loci.
Given that our aim was to study population genetic structure within the IT lineage, we first removed all individuals that showed any sign of introgression from other lineages (see Yang et al. 2018; Ruiz Miñano et al. 2022). To this end, we retained only the first SNP per RAD locus (reducing the data from 103,918 SNPs to 35,227 SNPs) and quantified the probability of being assigned to the IT lineage based on ADMIXTURE with K = 2. We subsequently removed all individuals whose probability of being assigned to the IT lineage was <0.99. This resulted in the retention of 415 individuals from 34 populations (Fig. 1), all belonging to the IT lineage, with an average of 12 (range 4–15; see Table S1) individuals genotyped per population. These individuals were subsequently subject to spatial analyses with distance-based Moran Eigenvector Maps (dbMEMs) and genotype-environment associations as described in detail below.
Climatic variables
We ran principal component analyses on 19 bioclimatic variables extracted from WorldClim 2.0 (Fick 2017) at a 30 arc-sec spatial (~1 km2) resolution for the 34 sampling locations analysed. The first three axes explained 90% (51.5, 20.4, and 17.7%, respectively) of the variation (Fig. 1 illustrates these with spatial interpolation, using the idw function in the rspatial package; Table S2). PC1 represents a gradient from hot and dry (high values) to cool and wet (low values) climate, and the geographic distribution of the scores clearly separates lowland regions from mountains (Fig. 1). High values for PC2 represent locations with a consistently high temperature across the year (i.e., mild winters and hot summers) and high precipitation seasonality (wet winters and dry summers), which results in a pronounced shift in PC2 from the southwest towards the northeast (Fig. 1). Values for PC3 capture variation that represents locations that are relatively cool and dry (high values) to warm and wet (low values; Fig. 1). These principal components adequately capture the Köppen–Geiger climate classification for Italy (Beck et al. 2018) and were used in all further analyses.
Spatial genetic structure
We investigated the spatial structure of genetic variation using the data set derived from a single SNP per RAD locus. We used multivariate regression models, redundancy analyses (RDAs), and distance-based Moran Eigenvector Maps (dbMEM). The methods are explained by Legendre and Legendre (2012), and all analyses were carried out in R using the package adespatial (Dray et al. 2021; see Borcard et al. 2018). Redundancy analyses are an extension of linear regression models for multivariate response variables and are suitable for the analyses of genomic data (Forester et al. 2018). dbMEM variables are the eigenvectors of a spatial matrix calculated from the geographical coordinates of the sampling locations (Dray et al. 2006; Legendre and Legendre 2012). These eigenvectors represent orthogonal spatial descriptors of the genetic variation across sampling locations at different spatial scales (defined by Moran’s I). dbMEMs variables can therefore be used as spatial explanatory variables in multiple regression or redundancy analysis of genetic variation.
We created a matrix of geographic distance between locations using the coordinates of each sampling location. Locations were linked within a minimum geographic distance using the longest edge in a minimum spanning tree (Fortin and Dale 2005). We calculated allele frequencies per population and transformed the data using Hellinger transformation (Borcard et al. 2018). This transformation consists of dividing each value in the data matrix by its row sum and taking the square root of the quotient, and thus gives low weight to variables with low counts or lots of zeros (i.e., rare alleles). Isolation by distance (spatial autocorrelation) is expected for population genetic data and this relationship should be removed before further analysis of dbMEMs (Legendre and Legendre 2012). We removed the linear trend (i.e., isolation by distance) by running a linear model of allele frequencies regressed on geographical coordinates and used the residuals for further analyses (Legendre and Legendre 2012). We subsequently ran a global RDA on the detrended population genetic data and used forward selection to identify significant dbMEMs (Blanchet et al. 2008). The significant dbMEMs were subsequently used as independent variables in the final RDA of the genetic data, resulting in one or more canonical axes that represent the spatially structured variation of population allele frequencies across the landscape. We then tested whether these canonical axes were associated with our climatic predictors (PC1, PC2, and PC3) using linear regressions. In addition, we performed a variance partitioning analysis (Legendre and Legendre 2012; Peres-Neto et al. 2006). This method is implemented in the vegan package (Oksanen et al. 2019) and uses partial redundancy analyses to quantify the unique and shared fractions of the genetic variation explained by the three main predictors: the linear (i.e., isolation by distance) and non-linear (i.e., dbMEMs) spatial structure, and climate (i.e., the three climatic PCs). Since the linear trend should be included in the variance partitioning, the data was not detrended before analysis (Legendre and Legendre 2012). Statistical significance and unique and shared contributions of isolation by distance, non-linear spatial structure, and climate were determined by ANOVA using 999 permutations (Borcard et al. 2018).
Genotype-environment associations
Analysis of genotype-environment association (GEA) aims to detect polygenic responses to selection by investigating changes in allele frequencies (Brauer et al. 2018; Forester et al. 2018). To identify outliers associated with the three climatic principal components, we filtered the 103,918 SNPs obtained after quality control (described above) for the 415 individuals with respect to linkage disequilibrium (LD). To ensure that our results were robust, we ran the subsequent analyses for two conservative cut-offs for LD: r2 = 0.8 and r2 = 0.5.
We used two different GEA approaches to identify SNPs associated to climate: latent factor models (LFMM; Frichot et al. 2013) and redundancy analyses (Bourret et al. 2014). Both LFMM and RDA assume linear relationships between genetic data and environmental variables (for a methodological review, see Forester et al. 2018). LFMM is a univariate approach that identifies associations between a single locus and environmental variables, using latent factors to correct for confounding effects. We used the lfmm package (Caye et al. 2019) to run latent factor models. As population structure is known to be an important confounding factor when detecting genotype associations (Caye et al. 2019), we first investigated the evidence for population structure using principal component analyses, and by estimating the number of ancestry components using a sparse non-negative matrix factorization algorithm (function snmf in the LEA package; Frichot and Francçois 2015). We tested K values ranging from one to ten, plotted the cross-entropy values, and selected K based on the inflection point. We calculated genomic inflation factor (GIF) and adjusted the false discovery rate using a Benjamini-Hochberg algorithm (François et al. 2016).
The approach based on RDA detects signatures of selection as a function of a multivariate set of predictors (Forester et al. 2018). Redundancy analyses were performed using the RDA function from the vegan package. For this method, we modelled allele frequency as a function of the three climatic PCs and produced three constrained axes (the same as the number of climatic predictors). We identified outlier loci on the constrained axes based on the “locus score”, which represents the loading of each locus in the ordination space. We calculated the mean locus score across all loci for each significant (i.e., p < 0.05) RDA axis, and individual loci with a score greater than 3.5 standard deviations from the mean were considered candidates for selection (Forester et al. 2016). Since the RDA approach is more powerful when not including factors that account for population structure (Forester et al. 2018), we did not include any such variable in the final model. We assessed multicollinearity between variables (climatic variables and dbMEMs) using the variation inflation factor (VIF); all VIF were <2 and therefore all were retained in the final model. All the statistical analyses were performed in R v. 4.0.2.
To infer the potential function for candidate genes, we conducted enrichment analyses based on Gene Ontology (GO) annotation using clusterProfiler package with default parameters (Yu et al. 2012) in R, for the outliers identified by RDA (which is the method considered to deliver the most robust list of candidates; Forester et al. 2018) and for the final outlier data set representing SNPs found by both LFMM and RDA approaches.
Results
Genetic differentiation across climatic regimes
After stringent filtering for sequencing depth and rate of missing data, and excluding all but the first SNP per RAD locus, 35,227 SNPs were retained. Expected heterozygosity, observed heterozygosity, and inbreeding coefficients were similar across populations (e.g., average expected heterozygosity, HE = 0.199, range 0.181–0.209; see Table S3), and uncorrelated with any of three climatic PCs (all p > 0.05). The global Fst for the 34 populations was 0.0615 (CI: 0.0606 – 0.0642 from 100 iterations of bootstrap).
The Hellinger transformed population allele frequencies showed a significant linear association with geographic coordinates (F2,31 = 2.92, P < 0.001; Fig. S1). The data were therefore detrended for the subsequent analysis of spatial structure using dbMEMs (Legendre and Legendre 2012). After forward selection, four dbMEMs describing population allele frequencies were retained (P ≤ 0.05; Table S4; Fig. S2, S3). A redundancy analysis on these four significant dbMEMs as response variables resulted in four canonical axes that together explained 6.0% of the variation (F4,29 = 1.52, P < 0.001; Fig. 2; Table 1). The first two canonical axes describe a rather broad-scale spatial structure in allele frequencies and the other two finer scale spatial structure (Fig. 2). None of these canonical axes were significantly associated with the climatic predictors (linear regressions with PC1, PC2, and PC3 as predictors; Table S5).
A RDA triplots of the Hellinger transformed allele frequency data. Dots represent sampling locations and arrows the four dbMEMs retained following forward selection. B dbMEM fitted scores for the four significant canonical axes of the redundancy analysis. Black dots represent positive values and white dots negative values. C Variance partitioning of the allele frequency data with unique and shared fractions of explained variation.
Variance partitioning analyses makes it possible to assess the unique and shared fraction of genomic variation explained by the three main predictors: the linear trend, spatial structure (i.e., four dbMEMs), and climate variables (i.e., three climatic PCs). The total amount of variation in the Hellinger transformed population allele frequencies explained by the linear trend, dbMEMs and climate was 17.6%. An RDA on the non-detrended (i.e., without removing linear trend) Hellinger transformed allele frequencies, using forward selection of climatic principal components, resulted in retention of climatic PC1 and PC2. These two predictors uniquely explained about 2% of the variation in allele frequencies (R2adj = 0.02, F 2,25 = 1.33, P = 0.002; Fig. 2). An additional 3% of variation explained by climate was collinear with the linear trend or dbMEMs. The spatial structure (linear trend and dbMEMs) alone accounted for a total of 15.1 % of variation in the Hellinger transformed genetic variation, with the unique proportions of variance being statistically significant for both the linear trend (R2adj = 0.091, F 2,25 = 2.49 P < 0.001; Fig. 2) and the non-linear spatial structure captured by the dbMEMs (R2adj = 0.047, F 4,25 = 1.41, P < 0.001; Fig. 2).
Genotype-environment associations
After filtering on LD at r2 = 0.8, we retained 80,285 SNPs for the analyses of genotype-environment associations. Using the more stringent filtering of r2 = 0.5 resulted in ~10% fewer SNPs (72,350), but a very high overlap in terms of significant outlier SNPs following LFMM and RDA (see below). This suggests that the results are robust at varying cut-offs, and we therefore report here only the results for the data set filtered at r2 = 0.8.
A principal component analysis and the broken stick method did not support a significant genetic clustering (Fig. S4), but sNMF identified six clusters (Fig. S5). We therefore set K = 6 in the LFMM analyses. The LFMM identified 221 candidate loci, of which 75 were associated with climatic PC1, 57 with PC2, and 89 with PC3. Manual adjustment of the genomic inflation factors (GIF) had only minor effect on these numbers.
The RDA returned three significant axes, on which we identified 820 unique candidate loci. The majority of these SNPs were most strongly associated with climatic PC1 (361), followed by PC3 (247) and PC2 (212). GO enrichment for these RDA outliers can be found in Table S6.
Out of these candidate SNPs, 48 were detected by both LFMM and RDA (the corresponding number for LD filtering at r2 = 0.5 was 43, of which 38 were shared with results for r2 = 0.8). These 48 outliers were dispersed throughout the genome, found on 16 of the 19 chromosomes; 23 outliers were most strongly associated with climatic PC1, 11 with PC2, and 14 with PC3. These SNPs were associated with 37 annotated genes (see Discussion and Table S7). The GO enrichment for these genes identified molecular functions related to catabolic processes (GO:0044248, GO:1901575, and GO:0009056), metabolic processes (GO:0044237 and GO:0008152), and oxidoreductase activity (GO:0016491).
Discussion
Quantifying the extent of climate-associated population genetic differentiation, and its spatial scale, is important for understanding past and future adaptation to climatic regimes. In this study, we investigated the relationship of genome-wide SNPs with climatic and spatial data across 34 populations of the common wall lizard (Podarcis muralis) in central Italy. This region is characterised by very high lizard densities, no physical barriers to gene flow, consistently high estimates of population genetic diversity (even at climatic extremes), and a genetic structure characterized by isolation by distance (Ruiz Miñano et al. 2021; Yang et al. 2018). Overall, our results demonstrate that climatic selection is sufficient to overcome high levels of gene flow and cause genetic differentiation, consistently sorting particular alleles across the landscape according to climatic regimes. As a result, we expect wall lizards to show pronounced local adaptation in morphology, physiology, and behaviour even across relatively small spatial scales.
In support of previous work (Yang et al. 2018), most of the genetic structure within central Italy, accounting for 12.9% of the total population genomic variation, represents a linear trend that is consistent with isolation by distance. The fine- to medium-scale spatial structure was less pronounced (uniquely explaining ~4.5% of population genetic variation), which is consistent with the lack of physical barriers to gene flow and absence of historical isolation of allopatric populations within the region (Yang et al. 2022). Excluding the additional variation explained by climate (see below), it is therefore perhaps not surprising that >80% of population genomic variation remained unexplained. Nevertheless, we cannot exclude that some of this unexplained variation is caused by historical or more recent gene flow from peripheral populations or lineages (Yang et al. 2022).
Our emphasis in this study was to test if the climatic variation within this region contributed to genome-wide patterns of genetic diversity and population differentiation. While there was no evidence that populations at climatic extremes have lower genetic variation, climate explained about 5% of the population genomic variation across the 34 populations. This result contributes to a growing literature on climate-associated genome-wide population differentiation in lizards (e.g., Rodríguez et al. 2017; Farleigh et al. 2021; for a study testing non-climate environmental influence on genome wide population differentiation of three species of lizard, see Krohn et al. 2019). While these studies do support a role for climate in shaping genetic differentiation, they are also somewhat limited by virtue of being largely descriptive. Further work would therefore benefit from sampling at spatial scales and across climatic regimes that allow more explicit tests of, for example, the diluting effect of gene flow on adaptive population divergence. Lineages or species that evolve along similar climatic gradients would also be interesting contrasts, in particular for identifying the extent to which climatic adaptation makes use of similar genes and pathways (e.g., Feiner et al. 2018).
About half of the genomic variation explained by climate was collinear with a linear shift in allele frequencies. This collinearity reflects the transition from a temperate, hot, and dry climate of the south-west coast, to a more oceanic climate with no dry season in the Apennine mountains and in the north-east of the distribution of the IT lineage of P. muralis (see Yang et al. 2022 for the phylogeography of this species). The two percent of genomic variation explained uniquely by climate represents fine- to medium-scale climatic variation not captured by either the linear or non-linear spatial structure. Common wall lizards are extremely abundant throughout central Italy and inhabit a wide range of microhabitats except for the hottest and driest environments (where the congener P. siculus entirely takes over; Capula et al. 1993). Future studies should test the extent to which the unexplained genetic variation could be accounted for by additional landscape features (e.g., habitat type, presence of competitors, human habitation; Beninde et al. 2016; Van Buskirk and Jansen van Rensburg 2020).
Common garden experiments and reciprocal transplants of lizards between climatic regimes have established that growth and other fitness proxies are commonly highest in the local environment (e.g., Niewiarowski and Roosenburg 1993; Uller and Olsson 2003; Iraeta et al 2006; While et al. 2015b). For example, reproductive biology and developmental rates can be locally adapted to the length of the activity season and the thermal conditions during spring and summer (reviewed in Uller and While 2015; Pettersen 2020). These traits provide a wide range of targets for selection that can contribute to population genetic differentiation. The ontology of genes associated with outlier SNPs is rather uninformative since the categories (e.g., metabolic activity) are broad and difficult to link to a priori expectations. However, a few of the most robust outlier SNPs (i.e., those identified by multiple methods) were associated with candidate genes that functional studies have shown to be important to thermal physiology. These include CDK4, a cyclin-dependent kinase, that is repressed in response to freezing temperatures in frogs (Zhang and Storey 2012), and a member of the heat shock protein family (HSPA8), which is a constitutively expressed chaperone that has been shown to have a seasonal expression pattern in heat- and cold-adapted goat breeds (Banerjee et al. 2014; Singh et al. 2017).
We may also expect to identify genes involved in regulation of development, metabolism, growth, and biological cycles, such as annual reproductive activity. The dual oxidase 2 (DUOX2) and its accessory protein (DUOXA2), both located on chromosome 14, are putative candidates since they are involved in thyroid hormone biosynthesis (Carvalho and Dupuy 2013). Thyroid hormone has been shown to mediate the effects of temperature on reptile reproductive physiology (Norris and Lopez 2011) and regulate embryonic growth and development (Ruuskanen and Hsu 2018). For example, experimental manipulation of thyroid hormones in ovo can modify the time to hatching in turtles and birds (e.g., McGlashan et al. 2017; reviewed in Ruuskanen and Hsu 2018). Thyroid hormone regulation is therefore likely to play important roles in climate adaptation in ectotherm vertebrates, including high developmental rate in cool-adapted populations (Pettersen 2020); an adaptation demonstrated to evolve rapidly in P. muralis (While et al. 2015b; Feiner et al. 2018).
It is not surprising to find candidate genes that are consistent with the expectation that climatic selection will lead to population divergence in genes that regulate thermal physiology and annual life cycles. In common wall lizards, climate is also associated with a marked shift in several morphological characters that are under sexual selection, including head size, shape, and colour ornamentation (While et al. 2015a; Ruiz Miñano et al. 2021). Theoretical studies suggest that exaggerated armaments and ornaments could be explained by an increased opportunity for female monopolization by dominant males in climates that support a long reproductive season and less synchronous female reproduction (reviewed in Shuster and Wade 2003; García-Roa et al. 2020). This adds another selective pressure that varies across the landscape in association with climate, and could contribute to divergence of genes that may at first appear of little relevance for climate adaptation per se. One of the candidate genes identified here, RAB7A, may reflect this climatic effect on sexual selection, and in particular sexual ornamentation. This gene encodes a Rab GTPase that regulates transport of early and intermediate melanosomes (Gomez et al. 2001; Fukuda 2021), the melanin-containing organelles of the melanophores in lizard skin (Taylor and Hadley 1970). RAB7A may also be involved in guanine synthesis or transport in iridophores (Stuckert et al. 2019), another cell type that can be responsible for the difference between brown and green skin in lizards (Eisentraut 1950; Kuriyama et al. 2017; Kuriyama et al. 2020). Thus, we hypothesize that this gene is involved in the expression of the conspicuous black and green colour ornamentation (i.e., “P. muralis nigriventris”; Böhme 1986) that is closely associated with coastal climate in P. muralis from western central Italy (Ruiz Miñano et al. 2021).
It is encouraging to find candidate genes that have previously been shown to be involved in both thermal physiology and animal colouration. However, genotype-environment associations are limited in the extent to which they can inform us about causality. Thus, these candidates remain putative loci of adaptation until quantitative experiments confirm the results. Furthermore, RAD data only covers a small part of the genome and most major effect candidates will therefore go undetected. Nevertheless, the results of this study indicates that climatic selection may affect a broad range of genes dispersed throughout the genome. Whole-genome sequences across steep climatic gradients, accompanied by transitions in phenotype (Ruiz Miñano et al. 2021), could help to further identify such genomic targets of climatic selection.
In summary, this study demonstrates that climate exercises an effect on population genetic differentiation in an abundant lizard with extensive gene flow. Further, the analyses suggest that particular alleles are consistently favoured in certain climatic regimes. More targeted approaches, combining functional studies with whole-genome sequencing, will be necessary to identify additional genes underlying population differentiation and local adaptation.
Data availability
All sequence data are available at GenBank, accession number: PRJNA486080. Population level data included as supplementary information (Table S1).
References
Andrade P, Pinho C, Pérez i de Lanuza G, Afonso S, Brejcha J, Rubin C-J et al. (2019) Regulatory changes in pterin and carotenoid genes underlie balanced color polymorphisms in the wall lizard. Proc Natl Acad Sci USA 201820320
Banerjee D, Upadhyay RC, Chaudhary UB, Kumar R, Singh S, Ashutosh et al. (2014) Seasonal variation in expression pattern of genes under HSP70. Cell Stress Chaperones 19:401–408
Bassitta M, Brown RP, Pérez-Cembranos A, Pérez-Mellado V, Castro JA, Picornell A et al. (2021) Genomic signatures of drift and selection driven by predation and human pressure in an insular lizard. Sci Rep 11:6136
Beck HE, Zimmermann NE, McVicar TR, Vergopolan N, Berg A, Wood EF (2018) Present and future Köppen–Geiger climate classification maps at 1-km resolution. Sci Data 5:180214
Beninde J, Feldmeier S, Werner M, Peroverde D, Schulte U, Hochkirch A et al. (2016) Cityscape genetics: structural vs. functional connectivity of an urban lizard population. Mol Ecol 25:4984–5000
Blanchet FG, Legendre P, Borcard D (2008) Modelling directional spatial processes in ecological data. Ecol Model 215:325–336
Böhme W (1986) Handbuch der Reptilien und Amphibien Europas. Aula-Verlag, Wiesbaden, Germany
Borcard D, Gillet F, Legendre P (2018) Numerical ecology with R. Springer Verlag.
Bourret V, Dionne M, Bernatchez L (2014) Detecting genotypic changes associated with selective mortality at sea in Atlantic salmon: polygenic multilocus analysis surpasses genome scan. Mol Ecol 23:4444–4457
Brauer CJ, Unmack PJ, Smith S, Bernatchez L, Beheregaray LB (2018) On the roles of landscape heterogeneity and environmental variation in determining population genomic structure in a dendritic system. Mol Ecol 27:3484–3497
Campbell-Staton SC, Edwards SV, Losos JB (2016) Climate-mediated adaptation after mainland colonization of an ancestrally subtropical island lizard, Anolis carolinensis. J Evol Biol 29:2168–2180
Campbell-Staton SC, Cheviron ZA, Rochette N, Catchen J, Losos JB, Edwards SV (2017) Winter storms drive rapid phenotypic, regulatory, and genomic shifts in the green anole lizard. Science 357:495–498
Campbell-Staton SC, Winchell KM, Rochette NC, Fredette J, Maayan I, Schweizer RM et al. (2020) Parallel selection on thermal physiology facilitates repeated adaptation of city lizards to urban heat islands. Nat Ecol Evol 4:652–658
Capula M, Luiselli L, Rugiero L (1993) Comparative ecology in sympatric Podarcis muralis and P. sicula (Reptilia: Lacertidae) from the historical centre of Rome: What about competition and niche segregation in an urban habitat? Boll di Zoologia 60:287–291
Carvalho DP, Dupuy C (2013) Role of the NADPH Oxidases DUOX and NOX4 in Thyroid Oxidative Stress. Eur Thyroid J 2:160–167
Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA (2013) Stacks: an analysis tool set for population genomics. Mol Ecol 22:3124–3140
Caye K, Jumentier B, Lepeule J, François O (2019) LFMM 2: fast and accurate inference of gene-environment associations in genome-wide studies. Mol Biol Evol 36:852–860
Chang CC, Chow CC, Tellier L, Vattikuti S, Purcell SM, Lee JJ (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:7
Clobert J, Massot M, Le Galliard J-F (2012). Multi-determinism in natal dispersal: the common lizard as a model system, pp 29-40 in J Clobert, M Baguette, TG Benton and JM Bullock (eds), Dispersal ecology and evolution. Oxford University Press
Dray S, Legendre P, Peres-Neto PR (2006) Spatial modelling: a comprehensive framework for principal coordinate analysis of neighbour matrices (PCNM). Ecol Model 196:483–493
Dray S, Bauman D, Blanchet G, Borcard D, Clappe S, Guenard G et al. (2021) adespatial: Multivariate Multiscale Spatial Analysis. R package version 0.3-8. https://www.CRANR-projectorg/package=adespatial
Eisentraut M (1950) Die Eidechsen der Spanish Mittelmeer Inseln. Akademie-Verlag, Berlin
Fan Z, Silva P, Gronau I, Wang S, Armero AS, Schweizer RM et al. (2016) Worldwide patterns of genomic variation and admixture in gray wolves. Genome Res 26:163–173
Farleigh K, Vladimirova SA, Blair C, Bracken JT, Koochekian N, Schield DR et al. (2021) The effects of climate and demographic history in shaping genomic variation across populations of the Desert Horned Lizard (Phrynosoma platyrhinos). Mol Ecol 30:4481–4496
Feiner N, Rago A, While GM, Uller T (2018) Signatures of selection in embryonic transcriptomes of lizards adapting in parallel to cool climate. Evolution 72:67–81
Fick SE (2017) WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int J Climatol 4315:4302–4315
Forester BR, Lasky JR, Wagner HH, Urban DL (2018) Comparing methods for detecting multilocus adaptation with multivariate genotype–environment associations. Mol Ecol 27:2215–2233
Forester BR, Jones MR, Joost S, Landguth EL, Lasky JR (2016) Detecting spatial genetic signatures of local adaptation in heterogeneous landscapes. Mol Ecol 25:104–120
Fortin M-J, Dale M (2005). Spatial analysis: a guide for ecologists. Cambridge University Press
François O, Martins H, Caye K, Schoville Sean D (2016) Controlling false discoveries in genome scans for selection. Mol Ecol 25:454–469
Fraser DJ, Weir LK, Bernatchez L, Hansen MM, Taylor EB (2011) Extent and scale of local adaptation in salmonid fishes: review and meta-analysis. Heredity 106:404–420
Frichot E, François O (2015) LEA: An R package for landscape and ecological association studies. Meth Ecol Evol 6:925–929
Frichot E, Schoville SD, Bouchard G, François O (2013) Testing for associations between loci and environmental gradients using latent factor mixed models. Mol Biol Evol 30:1687–1699
Fukuda M (2021) Rab GTPases: Key players in melanosome biogenesis, transport, and transfer. Pigment Cell Melanom Res 34:222–235
García-Roa R, Garcia-Gonzalez F, Noble DWA, Carazo P (2020) Temperature as a modulator of sexual selection. Biol Rev 95:1607–1629
Gibson MJS, Moyle LC (2020) Regional differences in the abiotic environment contribute to genomic divergence within a wild tomato species. Mol Ecol 29:2204–2217
Gomez PF, Luo D, Hirosaki K, Shinoda K, Yamashita T, Suzuki J-i et al. (2001) Identification of rab7 as a melanosome-associated protein involved in the intracellular transport of tyrosinase-related protein 1. J Invest Dermatol 117:81–90
Halbritter AH, Fior S, Keller I, Billeter R, Edwards PJ, Holderegger R et al. (2018) Trait differentiation and adaptation of plants along elevation gradients. J Evol Biol 31:784–800
Hargreaves AL, Germain RM, Bontrager M, Persi J, Angert AL (2020) Local adaptation to biotic interactions: a meta-analysis across latitudes. Am Nat 195:395–411
Horvathova T, Cooney CR, Fitze PS, Oksanen TA, Jelic D, Ghira I et al. (2013) Length of activity season drives geographic variation in body size of a widely distributed lizard. Ecol Evol 3:2424–2424
Iraeta P, Monasterio C, Salvador A, Diaz JA (2006) Mediterranean hatchling lizards grow faster at higher altitude: a reciprocal transplant experiment. Funct Ecol 20:865–872
Keller I, Alexander JM, Holderegger R, Edwards PJ (2013) Widespread phenotypic and genetic divergence along altitudinal gradients in animals. J Evol Biol 26:2527–2543
Krohn AR, Diepeveen ET, Bi K, Rosenblum EB (2019) Local adaptation does not lead to genome-wide differentiation in lava flow lizards. Ecol Evol 9:6810–6820
Kuriyama T, Esashi J, Hasegawa M (2017) Light reflection from crystal platelets in iridophores determines green or brown skin coloration in Takydromus lizards. Zoology 121:83–90
Kuriyama T, Murakami A, Brandley M, Hasegawa M (2020) Blue, black, and stripes: evolution and development of color production and pattern formation in lizards and snakes. Front. Ecol Evol 8:232
Legendre P, Legendre L (2012) Numerical ecology. Elsevier
MacGregor HEA, While GM, Barrett J, Guillem P (2017) Experimental contact zones reveal causes and targets of sexual selection in hybridizing lizards. Funct Ecol 31:742–752
Maruki T, Lynch M (2017) Genotype calling from population-genomic sequencing data. G3-Genes Genom Genet 7:1393–1404
McGlashan JK, Thompson MB, Van Dyke JU, Spencer RJ (2017) Thyroid hormones reduce incubation period without developmental or metabolic costs in Murray River short-necked turtles (Emydura macquarii). Physiol Biochem Zool 90:34–46
Niewiarowski PH, Roosenburg W (1993) Reciprocal transplant reveals sources of variation in growth rates of the lizard Sceloporus undulatus. Ecology 74:1992–2002
Norris DO, Lopez KH (2011) Hormones and reproduction of vertebrates, vol 3. Reptiles. Academic Press, Burlington, MA
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D et al. (2019). vegan: Community Ecology Package. R package version 2.5-6. https://CRAN.R-project.org/package=vegan
Olsson M, Shine R (2003) Female-biased natal and breeding dispersal in an alpine lizard, Niveoscincus microlepidotus. Biol J Linn Soc 79:277–283
Ortega J, Martín J, Crochet P-A, López P, Clobert J (2019) Seasonal and interpopulational phenotypic variation in morphology and sexual signals of Podarcis liolepis lizards. PLOS ONE 14:e0211686
Oufiero CE, Angilletta MJ (2006) Convergent evolution of embryonic growth and development in the eastern fence lizard (Sceloporus undulatus). Evolution 60:1066–1075
Peres-Neto PR, Legendre P, Dray S, Borcard D (2006) Variation partitioning of species data matrices. Ecology 87:2614–2625
Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE (2012) Double digest RADseq: an inexpensive methodfor de novo SNP discovery and genotyping in model and non-model species. PLoS One 7:e37135
Pettersen AK (2020) Countergradient variation in reptiles: thermal sensitivity of developmental and metabolic rates across locally adapted populations. Front Physiol 11:547
Prates I, Penna A, Rodrigues MT, Carnaval AC (2018) Local adaptation in mainland anole lizards: Integrating population history and genome–environment associations. Ecol Evol 8:11932–11944
Rodríguez A, Rusciano T, Hamilton R, Holmes L, Jordan D, Wollenberg Valero KC (2017) Genomic and phenotypic signatures of climate adaptation in an Anolis lizard. Ecol Evol 7:6390–6403
Ruiz Miñano M, While GM, Yang W, Burridge CP, Sacchi R, Zuffi M et al. (2021) Climate shapes the geographic distribution and introgressive spread of colour ornamentation in common wall lizards. Am Nat 198:379–393
Ruuskanen S, Hsu B-Y (2018) Maternal thyroid hormones: an unexplored mechanism underlying maternal effects in an ecological framework. Physiol Biochem Zool 91:904–916
Savolainen O, Lascoux M, Merilä J (2013) Ecological genomics of local adaptation. Nat Rev Genet 14:807–820
Schulte U (2008). Die Mauereidechse. Erfolgreich im Schlepptau des Menschen. Laurenti Verlag.
Shuster SM, Wade M (2003) Matings systems and strategies. Princeton University Press, Princeton, NJ
Sindaco R, Razzetti E, Doria G, Bernini F (2006). Atlas of Italian amphibians and reptiles. Edizioni Polistampa Firenze.
Singh KM, Singh S, Ganguly I, Nachiappan RK, Ganguly A, Venkataramanan R et al. (2017) Association of heat stress protein 90 and 70 gene polymorphism with adaptability traits in Indian sheep (Ovis aries). Cell Stress Chaperone 22:675–684
Stuckert AMM, Moore E, Coyle KP, Davison I, MacManes MD, Roberts R et al. (2019) Variation in pigmentation gene expression is associated with distinct aposematic color morphs in the poison frog Dendrobates auratus. BMC Evol Biol 19:85
Taylor JD, Hadley ME (1970) Chromatophores and color change in the lizard, Anolis carolinensis. Z für Zellforsch und Mikroskopische Anat 104:282–294
Telemeco RS, Radder RS, Baird TA, Shine R (2010) Thermal effects on reptile reproduction: adaptation and phenotypic plasticity in a montane lizard. Biol J Linn Soc 100:642–655
Tigano A, Friesen VL (2016) Genomics of local adaptation with gene flow. Mol Ecol 25:2144–2164
Uller T, Olsson M (2003) Life in the land of the midnight sun: are northern lizards adapted to longer days? Oikos 101:317–322
Van Buskirk J, Jansen van Rensburg A (2020) Relative importance of isolation-by-environment and other determinants of gene flow in an alpine amphibian. Evolution 74:962–978
Uller T, While GM (2015). Evolutionary ecology of reproductive investment in lizards. In: Rheubert JL, Siegel DS and Trauth SE (eds) Reproductive biology and phylogeny of lizards and Tuatara. CRC Press.
Warner DA, Shine R (2008) Determinants of dispersal distance in free-ranging juvenile lizards. Ethology 114:361–368
While GM, Williamson J, Prescott G, Horvathova T, Fresnillo B, Beeton NJ et al. (2015b) Adaptive responses to cool climate promotes persistence of a non-native lizard. Proc R Soc Lond B 282:1803
While GM, Michaelides S, Heathcote RJP, MacGregor HEA, Zajac N, Beninde J et al. (2015a) Sexual selection drives asymmetric introgression in wall lizards. Ecol Lett 18:1366–1375
Yang W, While GM, Laakkonen H, Sacchi R, Zuffi MAL, Scali S et al. (2018) Genomic evidence for asymmetric introgression by sexual selection in the common wall lizard. Mol Ecol 27:4213–4224
Yang W, Feiner N, Laakkonen H, Sacchi R, Zuffi MAL, Scali S et al. (2020) Spatial variation in gene flow across a hybrid zone reveals causes of reproductive isolation and asymmetric introgression in wall lizards. Evolution 74:1289–1300
Yang W, Feiner N, Salvi D, Laakkonen H, Jablonski D, While GM et al. (2022) Ancient lineage divergence, introgression and genomic diversity of common wall lizards (Podarcis muralis) reflect the dynamic history of the Mediterranean Basin. Mol Biol Evol, 39:msab311
Yeaman S (2015) Local adaptation by alleles of small effect. Am Nat 186:S74–S89
Yeaman S, Otto SP (2011) Establishment and maintenance of adaptive genetic divergence under migration, selection, and drift. Evolution 65:2123–2129
Yeaman S, Whitlock MC (2011) The genetic architecture of adaptation under migration-selection balance. Evolution 65:1897–1911
Yeaman S, Hodgins KA, Lotterhos KE, Suren H, Nadeau S, Degner JC et al. (2016) Convergent local adaptation to climate in distantly related conifers. Science 353:1431–1433
Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287
Zhang J, Storey KB (2012) Cell cycle regulation in the freeze tolerant wood frog, Rana sylvatica. Cell Cycle 11:1727–1742
Acknowledgements
We are grateful to Roberto Sacchi and Marco Zuffi for logistic support, Sozos Michaelides, Hannah MacGregor, Natalia Zajac, Ben Halliwell, Lindall Kidd, Rachel Lewandowsky, Hanna Laakkonen, Valentina Titone, and Nathalie Feiner for field assistance, Hanna Laakkonen for assistance in the molecular laboratory, Brenna Forester and Olivier François for statistical advice, and Nathalie Feiner for help with the preparation of figures. Three reviewers provided helpful feedback on the analyses and manuscript. The Ministry of Education, University and Research (MIUR) provided all the authorizations for the study, 2012–2013: Aut. Prot. PNM-0009344; 2014–2015: Aut. Prot. PNM-0011379; 2016–2018: Aut. Prot. PNM-0002154. This work was funded by the Royal Society of London (University Research Fellowship), the National Geographic Society, the British Ecological Society, the Swedish Research Council (2014-04465, 2017-03846), and a Wallenberg Academy Fellowship from the Knut and Alice Wallenberg Foundations (all to TU).
Funding
Open access funding provided by Lund University.
Author information
Authors and Affiliations
Contributions
MRM, GMW, and TU designed and coordinated the study; MRM, GMW, DS, and TU performed fieldwork and collected samples; WY coordinated, and MRM and WY performed, laboratory work; MRM, WY, and TU analysed data; MRM, GMW, WY, CPB, and TU interpreted the results. MRM and TU wrote the manuscript with comments from, and final version approved by, all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ruiz Miñano, M., While, G.M., Yang, W. et al. Population genetic differentiation and genomic signatures of adaptation to climate in an abundant lizard. Heredity 128, 271–278 (2022). https://doi.org/10.1038/s41437-022-00518-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-022-00518-0