Abstract
The warming climate will expose alpine species adapted to a highly seasonal, harsh environment to novel environmental conditions. A species can shift their distribution, acclimate, or adapt in response to a new climate. Alpine species have little suitable habitat to shift their distribution, and the limits of acclimation will likely be tested by climate change in the long-term. Adaptive genetic variation may provide the raw material for species to adapt to this changing environment. Here, we use a genomic approach to describe adaptive divergence in an alpine-obligate species, the white-tailed ptarmigan (Lagopus leucura), a species distributed from Alaska to New Mexico, across an environmentally variable geographic range. Previous work has identified genetic structure and morphological, behavioral, and physiological differences across the species’ range; however, those studies were unable to determine the degree to which adaptive divergence is correlated with local variation in environmental conditions. We used a genome-wide dataset generated from 95 white-tailed ptarmigan distributed throughout the species’ range and genotype–environment association analyses to identify the genetic signature and environmental drivers of local adaptation. We detected associations between multiple environmental gradients and candidate adaptive loci, suggesting ptarmigan populations may be locally adapted to the plant community composition, elevation, local climate, and to the seasonality of the environment. Overall, our results suggest there may be groups within the species’ range with genetic variation that could be essential for adapting to a changing climate and helpful in guiding conservation action.
Similar content being viewed by others
Introduction
Climate change is predicted to shift species distributions, changing the composition of biological communities (Parmesan and Yohe 2003), and testing the physiological tolerances of the species within them (Pörtner 2002). Species have multiple strategies at their disposal for responding to environmental change: moving to a more favorable location (Klanderud and Birks 2003), acclimating via phenotypic plasticity (Berteaux et al. 2004 [i.e., shifts in phenotypic distributions without underlying genetic change]), adapting to novel environments (Hoffmann and Sgró 2011), or some combination of all three (Bi et al. 2019). Range shifts are dependent on suitable habitat being within a reasonable dispersal distance (Thuiller 2004), and therefore are not always an effective response. In the long-term, the limits of a plastic phenotype are likely to be tested with climate change (Somero 2010; Laiolo and Obeso 2017). Adaptation, however, is contingent on there being sufficient standing genetic variation, originating from de novo mutation or immigration from other populations, for selection to act upon (Kawecki and Ebert 2004; Hoffmann and Sgró 2011). An understanding of the relationship between adaptive divergence and local environment underlies evolutionary potential and the species’ response to climate change and could ultimately provide insight into how to aid conservation planning.
One way to evaluate patterns of adaptive divergence in a species is to look for signals of selection (i.e., genetic loci consistent with expected patterns of natural selection) that are correlated with variables describing range-wide environmental variability. Genotype–environment association (GEA) analyses identify genetic loci under selection based on correlation with environmental variables that are hypothesized to drive natural selection (Coop et al. 2010; Rellstab et al. 2015; Forester et al. 2016). There are several advantages of using GEA analyses over the more traditional divergence-based approaches (e.g., Storz 2005) to identify loci under selection. First, GEA analyses provide a hypothetical environmental link to local adaptation that allows a broader ecological interpretation. However, careful consideration of the biological importance of evaluated variables is essential because many environmental variables are correlated and would, therefore, be indistinguishable in such an analysis. Second, multiple environmental variables may be involved in driving selection simultaneously. Recent application of multivariate constrained ordination techniques incorporates multiple environmental variables in a single analysis (Capblancq et al. 2018a; Forester et al. 2018). Finally, GEA analyses in general, and constrained ordination techniques in particular, are better at detecting relatively weak multi-locus selection than approaches based on genetic divergence alone (Brauer et al. 2016; Forester et al. 2018). Evaluating patterns of adaptive divergence may be particularly informative for species occupying habitats undergoing rapid environmental change due to shifting selection pressures likely to be encountered.
Alpine communities are among the most climate-threatened habitats in North America (Holsinger et al. 2019), having already seen significant declines that are expected to continue at a relatively fast pace (Pepin et al. 2015). The extreme environments of alpine communities are characterized by short growing seasons and an assemblage of plant and animal species evolved to thrive in these conditions (Rundel and Millar 2016). For animals, survival in an alpine community requires a combination of behavioral, morphological, energetic, and physiological traits (Martin and Wiebe 2004). As the climate warms, alpine species are expected to shift northward and/or to higher local elevation in search of suitable conditions (Freeman et al. 2018). However, alpine-adapted traits may result in a narrow niche breadth and a limited ability to respond to an altered environment (Laiolo and Obeso 2017; Resano-Mayor et al. 2019). Similarly, alpine species are often limited in their ability to move upward in elevation because they already occupy the highest elevations locally. But phenotypic plasticity and adaptation remain as potential responses to a warming climate. The pace and expected extremes of the warming at high elevations, suggest phenotypic plasticity may be a poor response to a changing environment in the long-term (Laiolo and Obeso 2017). The rapid rate of environmental change, and reduced plasticity/increased canalization of physiological/developmental adaptations may be a warning sign of more severe population declines of alpine specialists than of less specialized species. Previous work on alpine specialists have used genetic information to understand potential strategies for persistence in a changing climate (e.g., Henry et al. 2012). Similarly, GEA analyses may provide particularly important insight into the evolutionary capacity of alpine species by identifying the genetic signature and environmental drivers of local adaptation. Here, we focus on identifying spatially variable adaptive genetic variation in the white-tailed ptarmigan (Lagopus leucurus), an alpine-obligate bird (Martin et al. 2015) that is vulnerable to future changes in climate.
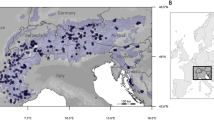
The white-tailed ptarmigan distribution extends across western North America, from the Yukon Territory, and Alaska, in the north to New Mexico in the south (Fig. 1). The entire life-cycle of the white-tailed ptarmigan is confined to climate-threatened alpine and sub-alpine habitat (Martin et al. 2015; Holsinger et al. 2019), indicating the species as a whole is threatened by climate change. In recent decades, populations of white-tailed ptarmigan have generally been stable in the few places where populations have been studied (Martin et al. 2004; Wann et al. 2014), although apparent local declines and high annual variation in population size have been observed (Martin et al. 2015; Wann et al. 2016), particularly at the southern extreme of the distribution in New Mexico (Braun and Williams 2015; New Mexico Department of Game and Fish 2016). At present there are five recognized subspecies of the white-tailed ptarmigan: L. l. peninsularis [“Kenai” hereafter], L. l. leucura [“Northern” hereafter], L. l. saxatilis [“Vancouver” hereafter], L. l. rainierensis [“Rainier” hereafter], and L. l. altipetens ([“Southern” hereafter]; Martin et al. 2015). The Vancouver (Martin et al. 2015) and Southern subspecies (United States Fish and Wildlife Service 2012), either have been or currently are being considered for formal protection resulting from vulnerability to climate change. Major breaks in genetic structure have been detected for the Vancouver and Southern subspecies. More subtle subdivision was identified within the Southern subspecies (Langin et al. 2018). In addition, differences in body size (Wilson and Martin 2011; Martin et al. 2015), gut length (Moss 1983), plumage (Martin et al. 2015), and bill morphology (Martin et al. 2015) have been observed across the species range. The north–south species distribution also coincides with an elevation gradient, where alpine habitat is at lower elevations at higher latitudes. Generally, the northern extent has relatively more shrubland than grassland, more snow, cooler summer temperatures, more annual variation in day length, and a later and shorter growing season (Table 1). There is interspecific competition among white-tailed, rock (L. muta), and willow (L. lagopus) ptarmigan (Hannon et al. 1998; Montgomerie and Holder 2008) in the northern part of the range that may have led to dietary specialization (Weeden 1967; Moss 1974, 1983; Bryant and Kuropat 1980) and/or niche partitioning (May and Braun 1972; Wilson and Martin 2008). Competition among congeners, observed differences in morphology, genetic structure, and local environment together suggest the potential for local adaptation. At present, no studies have characterized the relationship between adaptive divergence and environmental gradients, a goal that can be accomplished with a GEA analysis.
White-tailed ptarmigan distribution (a) with the approximate geographic boundaries of currently recognized subspecies (Martin et al. 2015) and sample locations. Individual states (U.S.), provinces (Canada), and regions (i.e., Vancouver Island) are labeled. White-tailed ptarmigan neighbor-joining trees based on proportion nucleotide differences (Nei and Kumar 2000) for all SNPs range-wide (b; color coding matches panel a), and within the Southern subspecies (c; dark blue = northern Colorado, light blue = central Colorado, green = southern Colorado, yellow = New Mexico) showing overall divergence patterns. Branches with support from <700 boostraps were collapsed.
Here we investigate the potential for environmental-associated local adaptation range-wide and among populations of the Southern subspecies. We extend previous work on range-wide variation by linking signals of selection to hypothetical drivers of selection (i.e., correlation to environmental variables) and, in some cases, potential gene function (i.e., pathway and functional annotation databases). The aim of our study was to shed light on how adaptive divergence may influence the ability of white-tailed ptarmigan to respond to climate change and provide additional insight into the genetic distinctness of subspecies of conservation concern.
Materials and methods
Study area and genetic data
We used previously generated genome-wide single nucleotide polymorphism (SNP) data composed of 14,866 loci, for 95 individuals from across the species’ natural range (Langin et al. 2018), geographically distributed as follows: Colorado (N = 35), New Mexico (N = 4), Washington (N = 12), Montana (N = 3), Alberta (N = 5), British Columbia (N = 6 mainland; N = 12 Vancouver Island), the Yukon (N = 6), and Alaska (N = 12; Fig. 1). The data were originally generated with a modified version (Langin et al. 2018) of the double-digest RADseq protocol (Peterson et al. 2012). For the current study, we estimated the order and genomic neighborhood of these de novo SNPs by alignment to the annotated genome of Meleagris gallopavo, or turkey (Dalloul et al. 2010 [GCA_000146605.1, UMD2.86 gene annotation version]), which as a new-world galliform is more closely related to L. leucura than is chicken (Persons et al. 2016). We used MegaBLAST (Morgulis et al. 2008) to map 88-bp consensus sequence tags generated from the ptarmigan samples with the ddRADseq approach (Langin et al. 2018), keeping only the tags that uniquely aligned or for which all secondary alignments were 30 bases or shorter. To ensure a high rate of correctly inferred orthology, the query coverage of the alignment was required to be nearly complete after allowing for small indels (88 ± 5 nt) and the percent of the alignment was required to be ≥90%. Of the original 14,866 SNPs, 8245 were aligned to 29 chromosomes and 47 unplaced scaffolds of the turkey genome for the present analysis. Mapping SNPs to the turkey genome allowed us to estimate linkage disequilibrium (LD) and identify gene annotations that are likely in LD with SNPs.
Linkage disequilibrium
With the 8245 SNPs aligned to the turkey genome we used all individuals to estimate LD decay based on turkey chromosome position to identify the distance at which candidate loci could be considered physically linked to a putative gene region on the reference genome. We phased our SNPs using BEAGLE 5.0, assuming a constant recombination rate of 1 cM per Mb (Browning and Browning 2007). We then calculated LD in vcftools (-hap-r2 command [Danecek et al. 2011]) at multiple distances (10 bp–1 Mbp). We considered SNPs with an LD of r2 ≥ 0.10 to be physically linked, a value consistent with previous studies (Vos et al. 2017).
Genotype–environment associations
Environmental covariates
We chose environmental covariates known to be important to white-tailed ptarmigan life-history characteristics to test for associations with allele frequency differences. We first checked variables for correlation within general categories (snow condition, climate, phenology, wind, topography, and vegetation), eliminating all but one highly correlated variable (Pearson’s r > |0.60|). Retained variables were subsequently evaluated for correlation, eliminating highly correlated variables based on our understanding of biology and the level of relative support in the literature; variables with more relative support and/or with more direct links to fitness impacts were prioritized. For example, winter precipitation was linked to white-tailed ptarmigan fitness consequences (Wann et al. 2016); therefore, all correlated variables were removed when lacking a documented impact of similar potential evolutionary importance. Several considered variables were highly correlated and would therefore be indistinguishable in our interpretation of potential drivers of selection. See Appendix A and Table B.1 for details on covariate calculation, correlation, and selection.
Snow condition
White-tailed ptarmigan use snow patches to regulate body temperature during summer and winter periods (Braun et al. 1976; Stokken 1993). Persistent snow can decrease breeding success (Clarke and Johnson 1992; Martin and Wiebe 2004), shift hatch date (Giesen et al. 1980), influence food accessibility, and ultimately seasonal movements (May and Braun 1972; Hakkarainen et al. 2007). We evaluated five snow cover (MODIS; Hall et al. 2006) and depth (Canadian Meteorological Centre; Brown and Brasnett 2010) variables: number of winter days with snow cover, snow melt rate, July snow cover, November snow cover, and winter snow depth. Northern locations generally had deeper snow, which persisted later into the summer (Table 1).
Climate
Precipitation can impact breeding phenology (Wann et al. 2016), demographic rates, and recruitment (Wann et al. 2014, 2016). Temperature can influence behavior, physiology and development (Martin et al. 2015), population growth rates (Wang et al. 2002a; Wann et al. 2016), breeding phenology (Wang et al. 2002b; Wann et al. 2016), and indirectly through effects on snow quality and winter roost site suitability (Braun et al. 1976). We evaluated six climate (WorldClim; Fick and Hijmans 2017) variables: July precipitation, winter precipitation, winter solar radiation, summer average temperature, summer precipitation, and summer maximum temperature. In general, the Southern subspecies experienced higher summer precipitation and temperatures than the Northern subspecies; however, Vancouver Island experienced the highest winter precipitation by far (Table 1).
Phenology
Short alpine growing seasons reinforce synchronization between the timing of resource availability and breeding phenology for the white-tailed ptarmigan (Giesen et al. 1980; Wann et al. 2016, 2019). Mismatch between habitat phenology and breeding phenology can impact demographic rates (Wann et al. 2014, 2016), forage quality (Wann et al. 2019), and chick survival (Wann et al. 2019). Timing of the availability of high-quality food resources could also impact the ability of the species to maintain their nearly continuous molt (Pyle 2007). Range-wide temporal variation in snow cover may result in variable selective pressures (Zimova et al. 2018) on a species with a complete color change molt (winter = white, summer = brown; Pyle 2007) that provides seasonal concealment (Hoffman and Braun 1975; Braun et al. 1976). Warming springs may result in early snow melt and increased predation risk due to camouflage mismatch (Imperio et al. 2013; Zimova et al. 2018). We evaluated three phenology variables: green-up (MODIS; Friedl et al. 2009), growing degree days, and a seasonality index (Daymet; Thornton et al. 1997, 2018). Generally, the Northern subspecies experienced a later start to the growing season and a lower seasonality index (Table 1).
Wind
Wind is a prominent component in alpine communities, essential for sweeping away snow and uncovering areas of vegetation for foraging (May and Braun 1972), and nest site selection (Giesen et al. 1980), as well as piling up snow for roosting (Braun et al. 1976). We evaluated two wind speed variables (Fick and Hijmans 2017; NOAA National Centers for Environmental Prediction (NCEP) 2018): winter coarse wind speed, and winter meridional wind speed. The Southern subspecies generally experienced higher wind speeds, although Vancouver Island at intermediate latitudes had the highest meridional wind speeds (Table 1).
Topography
Local topography in alpine communities can indirectly influence ptarmigan elevational migration (Hoffman and Braun 1975), dispersal (Fedy et al. 2008), nest site selection (Giesen et al. 1980; Spear et al. 2019), nest success (Wiebe and Martin 1998), and thermoregulation (Frederick and Gutiérrez 1992). The terrain (i.e., slope, aspect) may also drive selection through impacts on the quality and availability of local resources (i.e., forage, nesting sites, roosting sites). For example, northern orientation may avoid solar radiation, and facilitate snow persistence into summer months. We evaluated elevation, aspect, and slope (United States Geological Survey EROS Center 2007). The Northern subspecies occupied areas with relatively lower aspects and slopes than the Southern subspecies (Table 1). Notably, Vancouver Island corresponded to the highest slopes and most western aspects.
Vegetation
The composition of forage shrubs is known to vary throughout the range of the white-tailed ptarmigan (Weeden 1967; May and Braun 1972; Moss 1973) and correspond with physical digestive tract differences in other closely related tetraonids (Moss 1974, 1983), likely a result of different shrub species being differentially digestible (Bryant and Kuropa 1980; Palo 1984). Composition of vegetation can influence nest site selection (Spear et al. 2019), and demographic rates (Fedy and Martin 2011). We evaluated six land cover classes: sub-polar conifer forest, broad-leaf forest, mixed forest, grassland, grass-lichen-moss, and wetland (Latifovic et al. 2017). The northern subspecies were in areas with higher shrubland and forest cover and lower grassland cover than the Southern subspecies (Table 1). The exception was Vancouver Island which had the highest forest cover.
Genotype–environment associations
We used an individual-based redundancy analysis (RDA) implemented in the R package vegan (Oksanen et al. 2017) to identify candidate adaptive loci in two separate analyses: range-wide (N = 95) and within the Southern subspecies (N = 39). RDA combines multivariate linear regression and principal components analysis (PCA) to identify allele frequencies correlated with environmental covariates. Spearman rank correlation between dissimilarity measures and the environmental variables was highest with a Euclidean distance, thus use of RDA is appropriate (data not included, [Legendre and Legendre 2012]). RDA performs well relative to other GEA and divergence-based (i.e., without covariates) approaches (Capblancq et al. 2018b; Forester et al. 2018). In the range-wide analysis we accounted for spatial genetic structure by inclusion of Moran eigenvector maps (MEM) based on a Gabriel graph neighborhood and inverse distance weights using R packages spdep (Bivand and Piras 2015) and adespatial (Dray et al. 2019). Significant MEMs (i.e., those that explain spatial genetic structure) were identified in a forward selection algorithm (rda and ordistep, PIN = 0.01 in vegan) including all MEMs and the significant genetic axes (broken-stick criterion [Legendre and Legendre 2012]) of a principal coordinates analysis (pcoa in R package ape [Paradis and Schlier 2018]) on a Bray–Curtis genetic distance matrix (vegdist in vegan). The Bray–Curtis genetic distance allowed us to account for non-Euclidean spatial genetic structure. Our partial RDA included seven significant MEMs and environmental covariates for green-up, seasonality index, sub-polar conifer forest (conifer), broad-leaf forest, mixed forest, grassland, grass-lichen-moss, wetland, snow cover (November and July), winter days with snow, snow melt rate, winter snow depth, aspect, slope, summer maximum temperature, winter and July precipitation after accounting for pair-wise correlation (Pearson’s r ≥ |0.60|) and multicollinearity in the full model (VIF ≥ 10). Covariate means and standard deviations by recognized subspecies are included in Table 1. Loci in the tails of the SNP loading distribution for each significant (inspection of screeplot in Fig. B.1A; P < 0.01 from anova.cca with 999 permutations in vegan) marginal constrained RDA axis were considered candidate adaptive loci. We used a two-tailed P value based on 3 standard deviations as a cut off for candidacy (P < 0.003).
We identified candidate adaptive loci within the Southern subspecies as described above (see Fig. B.1B for scree plot). However, we did not include spatial predictors (i.e., MEMs) because genetic structure was low (global FST = 0.07), no genetic axes were significant by the broken-stick criterion (results not included), and controlling for low population structure can inflate false detections (Forester et al. 2018; Price et al. 2020). For comparison we include results including MEMs in the Appendix (Fig. B.2). Vegetation variables were combined into structural groups (i.e., forest, shrubland, grassland, wetland, barren) because variation within finer divisions across the southern range was low. Variables evaluated in the Southern subspecies RDA included forest, shrubland, grassland, wetland, July snow cover, winter days with snow, snow melt rate, green-up, seasonality index, aspect, slope, summer precipitation, summer maximum temperature, winter minimum temperature, winter coarse, and spring fine scale wind speed. Covariate means and standard deviations by genetic group within the Southern subspecies are included in Table 2.
Putative preliminary functional links
The majority of SNPs obtained with a reduced representation approach are intergenic and would be inherited in DNA blocks (i.e., in linkage). While we may infrequently sample a gene directly, we can expect that sampling a SNP linked to a gene under selection will produce the same signal as sampling the gene would. Estimation of LD decay can provide an indication of how distant we should expect linked SNPs and genes to be. However, LD decay varies across a genome (Slatkin 2008) and estimates can be inflated when multiple individuals are used in calculations (Mangin et al. 2012) as in the value we report here. We therefore chose a threshold for linkage much lower than our empirical LD decay (see Results section) that will result in a reasonable, conservative gene set associated with our candidate SNPs. For candidate SNPs (i.e., identified by the GEA) that aligned to the turkey genome using MegaBLAST, we used SnpEff (Cingolani et al. 2012) to identify genes within 5 kbp of, and therefore in linkage with, a candidate SNP. With the list of the genes identified as linked (<5 kbp) to a candidate SNP, we used default settings in DAVID (Database for Annotation, Visualization and Integrated Discovery; Huang et al. 2007) to identify enriched molecular pathways, functional annotation, and/or gene ontology (GO) terms.
Clustering
We created neighbor-joining trees to visualize patterns of differentiation, range-wide and within the Southern subspecies, for multiple SNP sets: autosomal candidate adaptive SNPs (candidate SNPs that did not map to turkey sex chromosomes), and putatively neutral SNPs. Neighbor-joining trees were constructed with the bionj function in R package ape, using an individual-based distance matrix calculated as the proportion of different nucleotide sites (Nei and Kumar 2000), excluding missing data in pair-wise estimations, and using 1000 bootstraps. Branches with support from <700 bootstraps were collapsed for visualization. We used a principal coordinates analysis (pcoa in R package ape) to visualize clustering of GO term-associated SNPs due to low sample size (N = 5 SNPs).
Results
Linkage disequilibrium
LD decayed to r2 < 0.10 at ~100 kbp (Fig. B.3). SNPs mapped to the turkey genome at a density of ~1 SNP per 168 kbp; smaller chromosomes generally had fewer SNPs. Our GO and pathway enrichment tests considered only annotated turkey genes within 5 kbp of the orthologous SNP coordinate of each aligned SNP. Actual genomic distances to any homologous gene of white-tailed ptarmigan are unknown. This operational threshold is much less than the average distance at which sites are empirically unlinked. We can therefore assume genes within this threshold distance are in aggregate an informative and conservative set for functional enrichment tests. Importantly, this approach does not make any assumption about LD beyond this threshold and does not preclude the possibility that selection on more distant gene features impact observed associations.
Genotype–environment associations
Spatial predictors (MEMs) accounted for 20% of the range-wide variance, while environmental variables accounted for 26%; 54% remained unaccounted for (adjusted r2 = 0.13). A total of 224 SNPs were in the distribution tails of the first three partial RDA axes, and therefore considered candidate adaptive loci: 48 on RDA1, 93 on RDA2, and 83 on RDA3 (see Table 1 for variable loadings and associated count of candidate loci). RDA1 separated Southern and Kenai on nearly opposite ends of an environmental gradient primarily of grassland cover (0.66), green-up (−0.35), winter snow depth (−0.38), November snow cover (−0.33), and July precipitation (0.35; Fig. 2a, Table 1). Southern individuals associated with higher grass cover, more July precipitation, and early green-up. RDA2 separated along an environmental gradient primarily of winter precipitation (Fig. 2a, b, Table 1; 70 candidate SNPs; loading = 0.66): Vancouver on the positive end (high winter precipitation), Rainier, Southern, and Northern approximately intermediate, and Kenai on the negative end. RDA3 primarily separates Kenai and Northern individuals (Fig. 2b) along an environmental gradient of forest cover (conifer forest [−0.46], broad-leaf forest [0.34]), snow (winter days with snow [−0.33]), and green-up (0.30). The Kenai subspecies was associated with more deciduous forest, less persistent snow, and later green-up.
Within the Southern subspecies, environmental variables accounted for 48% of the variance, and 52% remained unexplained (adjusted r2 = 0.10; no spatial predictors were included in this analysis). Seventy-nine candidate SNPs were identified on the first two axes: 38 on RDA1, 48 on RDA2 (see Table 2 for variable loadings and associated count of candidate loci). RDA1 separated birds from southern Colorado on one end and New Mexico on the other, along an environmental gradient of climate, wind, and vegetation variables (Table 2, Fig. 3). New Mexico has warmer winters and wetter summers relative to southern Colorado. RDA2 separates north-central Colorado birds from southern Colorado and New Mexico primarily along a gradient of summer precipitation and temperature (brood rearing precipitation [0.83], early summer maximum temperature [0.59], July snow cover [0.48], winter days with snow cover [0.41]). New Mexico and southern Colorado have warmer, wetter summers relative to the rest of Colorado (Table 2).
Putative preliminary functional links
SnpEff identified 32 genes on 19 chromosomes, associated with 37 annotations within 5 kbp of a candidate SNP; all 37 SNPs were intergenic (Table B.2). Enrichment tests in DAVID identified ten significantly overrepresented terms (P < 0.05), none of which remained significantly overrepresented after adjustment for multiple testing (Benjamini < 0.05). Significant categories were associated with either GO terms for Epidermal Growth Factor (EGF)-like domains or activin signaling pathways (Table B.3).
Candidate SNPs identified within the Southern subspecies were tightly linked to 12 annotated genes on 11 chromosomes. All candidate SNPs were intergenic (Table B.4). No pathways or GO terms were overrepresented based on this gene list.
Clustering
Range-wide, putatively neutral SNPs clustered individuals largely by recognized subspecies (Fig. 4a). Some individuals sampled within the described range of one subspecies clustered with individuals in a different subspecies. These deviations occurred among the Northern, Kenai, and Rainier subspecies where sampling density was generally low (see Figs. B.4 and B.5 for more details on sampling location and clustering). Autosomal candidate adaptive loci echo clustering of neutral loci (Fig. 4b).
Neighbor-joining trees based on proportion nucleotide differences (Nei and Kumar 2000) for different single nucleotide polymorphism (SNP) sets: range-wide putatively neutral (a; N = 14,642) and autosomal candidate adaptive (b; N = 213). Clustering based on SNPs associated with significant GO terms: Epidermal Growth Factor (EGF)-like domain (c; N = 5) is illustrated with the first two principal coordinate (PCoA) axes. Points correspond to individuals color coded by currently recognized subspecies or population. Symbols other than circles (■ or ▲) highlight subdivision or less defined boundaries within subspecies. Support from 1000 bootstraps shown in Figs. B.6 and B.7. Branches with support from <700 boostraps were collapsed.
Within the Southern subspecies, putatively neutral loci group New Mexico and southern Colorado birds into tight groups within the central and northern Colorado individuals (Fig. 5a). Autosomal candidate adaptive loci resulted in a clear separation of north-central Colorado, from both New Mexico and southern Colorado (Fig. 5b). At both scales, neighbor-joining trees with candidate adaptive loci resulted in longer branch lengths and more distinct groups. This was particularly true for Vancouver (Fig. 4b), New Mexico, and southern Colorado (Fig. 5b).
White-tailed ptarmigan Southern subspecies divergence patterns. Neighbor-joining trees based on proportion nucleotide differences (Nei and Kumar 2000) for different single nucleotide polymorphism (SNP) sets: for the Southern subspecies (39 individuals) putatively neutral SNPs (a; N = 10,946) and autosomal candidate adaptive SNPs (b; N = 80). Support from 1000 bootstraps shown in Figs. B.8and B.9. Branches with support from <700 boostraps were collapsed.
Discussion
Characterization of environmentally linked adaptive divergence can be used in conservation and management as additional information for conservation unit delineations, and/or understanding the potential for evolutionary capacity, particularly in response to climate change (Singh 2008). Overall, observed patterns of white-tailed ptarmigan divergence were similar to the previously identified strong divergence (neutral and putatively adaptive) of the Vancouver and Southern subspecies, reinforcing that boundaries for other subspecies could be further refined (Langin et al. 2018). The connections between the genome-wide pattern of adaptive divergence and environmental variables we found suggest there may be local adaptation to multiple variables likely to be impacted by climate change. We also found our candidate adaptive SNP set was enriched for a functional group (EGF-like domains) known to mediate a wide variety of protein–protein interactions and signaling pathways (Appella et al. 1988). Some of the functions attributed to this group are of particular relevance to white-tailed ptarmigan, climate change impacts, and conservation: adaptation to altitude (Ke et al. 2017; Xin et al. 2019) and latitude (Fabian et al. 2012), and seasonal pigment changing (Walker et al. 2007; Walker and Gunn 2010).
Descriptions of the local vegetation were the primary environmental variables associated with range-wide adaptive divergence (Table 1). These variables (primarily grassland cover) were highly correlated with elevation and latitude (Table B.5, Fig. 2a). Consequently, the main adaptive divergence gradient corresponds to a north–south, high-low latitude, low-high elevation, and approximately low-high grassland cover gradient. Our knowledge of white-tailed ptarmigan biology and life history suggests there are more than one possibly interacting, selection mechanisms that might be associated with local vegetation, elevation, and/or latitude. First, there may be local dietary adaptation. White-tailed ptarmigan in the Southern subspecies rely on willow (Salix spp.) for winter diet (May and Braun 1972). In the northern portion of the range, winter diet is more variable, including species of birch (Betula spp.) and alder (Alnus spp.) (Weeden 1967). Northern populations also have a larger gut size (Moss 1974) capable of acquiring nutrients from a more variable and lower quality diet (Weeden 1967; Bryant and Kuropat 1980). Dietary adaptation is not uncommon among alpine specialists (Rolando et al. 1997; Kohl et al. 2018), and would impact the potential for natural range shifts for the white-tailed ptarmigan. Given the Southern subspecies has the less variable diet, and birds in the northern part of the range appear to occupy more marginal alpine habitat (and consume a more variable diet) as a result of niche partitioning with congeners (Wilson and Martin 2008), the ability of the Southern subspecies to shift northward as the climate changes would likely be impacted by any potential dietary adaptation.
Second, white-tailed ptarmigan may be locally adapted to oxygen levels. Alpine plant communities at lower latitude are at higher elevations. Local adaptation to hypoxic conditions (like those at high elevation) has been described in multiple alpine specialists (Xin et al. 2019; Werhahn et al. 2020). Previous work suggests differential physiological responses to low oxygen environments in ptarmigan are primarily controlled by phenotypic plasticity (Carey and Martin 1997; Dragon et al. 1999). Our environmental gradients and tentative links to enriched functional groups (Fabian et al. 2012; Ke et al. 2017; Xin et al. 2019), however, indicate there may be adaptive divergence among populations of white-tailed ptarmigan to varying conditions along elevational and/or latitudinal gradients. Morphometric and life-history strategy differences between high latitude, low elevation populations (i.e., Yukon) and low latitude, high elevation populations (i.e., Colorado) further support adaptive divergence along these gradients (Wilson and Martin 2011). Natural dispersal, and transfer of genetic information, from the Southern subspecies northward would generally be inhibited by the large gaps in the alpine sky islands (Rundel and Millar 2016). Adaptive divergence associated with elevation and/or latitude would impose additional resistance to the ability of the Southern subspecies to shift down in elevation and up in latitude, as well as low elevation populations (Kenai and Vancouver) to shift range to higher latitudes and upslope.
We identified measures of phenology and snow melt on all three range-wide environmental selection gradients (Table 1), suggesting populations of white-tailed ptarmigan may be locally adapted to different local phenologies. Generally, the northern part of the white-tailed ptarmigan range experiences later snow melt, a later start to the growing season, and a different photoperiod than the southern part of the range. For many alpine and alpine breeding species, climate driven shifts in phenology have resulted in reproductive and molting mismatch (McKinnon et al. 2012; Pedersen et al. 2017). Earlier springs have led to earlier nest initiation and brood production in ptarmigan (García-González et al. 2016; Wann et al. 2016), which can lead to reduced fecundity when reproductive timing is out of synchrony with peak availability of plant and insect food (Wann et al. 2019).
In addition to the environmental variables, our tentative links to an enriched functional group suggests a connection between patterns of adaptive divergence and seasonal coat/plumage color change, based on findings for other species (Walker et al. 2007; Walker and Gunn 2010; Swanson et al. 2014). Ptarmigan molt into completely white plumage in the winter (Pyle 2007). Photoperiod is a well-known signal for molt and coat color change initiation in mammals (Zimova et al. 2018). Though the controls are less well understood in birds, hormones and snow melt timing likely play a significant role in addition to photoperiod (Höst 1942; Zimova et al. 2018). Plasticity in timing of molting in several species is considered limited (Charmantier et al. 2008, Zimova et al. 2018). Populations of the genus Lagopus that do not experience significant or prolonged snow may undergo only a partial color-transformation molt (Salomonsen 1936; Hannon et al. 1998; Jacobsen et al. 2007; Kozma et al. 2019), presumably due to relaxed selection pressure for completely white molt, suggesting genetic control of molt initiation. The amount of variation in the white-tailed ptarmigan for molting phenology is unknown. While no known populations are exempt from seasonal color change, initiation of nuptial molt may vary among populations in Colorado experiencing the same photoperiod (personal observation SJZ, CLA, GTW). The degree to which genetic variation underlies initiation of color change molt in white-tailed ptarmigan will impact the ability of the species to shift northward and respond plastically to changes in photoperiod, as well as to adapt as snow melt occurs earlier and seasonal timing changes.
Temperature and precipitation gradients were important components of environmental selection gradients range-wide (particularly winter precipitation) but were by far the largest components of environmental selection gradients within the Southern subspecies (Table 2). Observed adaptive divergence patterns associated with summer and winter (range-wide only) precipitation and temperature (Tables 1 and 2) are further supported as potential selective pressures with previous work in Colorado linking negative fitness consequences to wet and cold summers (Erikstad and Andersen 1983; Wann et al. 2016) and population growth and recruitment positively associated with winter precipitation (Wann et al. 2014). Subdivision within the Southern subspecies (Langin et al. 2018 (Fig. 5b here)) is of particular interest to conservation of the species, occurring on the extreme edge of the climate conditions expected to be most impacted by climate change. In addition to maintenance of potential locally adapted genetic variation being important for conservation of the currently occupied extent, genetic variation locally adapted to a warmer climate may be of particular value to conserving other portions of the white-tailed ptarmigan range if assisted migration or translocation are potential conservation actions. Signals of selection within the Southern subspecies identified New Mexico and southern Colorado birds as distinct from the north-central Colorado birds (and each other) along two environmental gradients: the first of winter temperature and summer precipitation, and the second of summer precipitation and summer temperature (Fig. 3 and Table 2). These populations may be important sources of genetic variation for the northern populations and their potential to adapt to warming local temperatures. Similarly, divergence of Vancouver along a primarily winter precipitation gradient suggests an additional source of genetic variation for higher winter precipitation (Fig. 2a, b).
Evidence of local adaptation in the New Mexico population is complicated by the re-introduction of 43 white-tailed ptarmigan individuals into the Pecos Wilderness area of New Mexico from northern Colorado in 1981 (Braun et al. 2011; Braun and Williams 2015). The New Mexico samples included in our analyses were collected from the Pecos Wilderness area. The re-introduced population in New Mexico displaying patterns of adaptive divergence distinct from the source population (northern Colorado) could be the result of high gene flow and transfer of beneficial alleles from southern Colorado, introgression from a remnant New Mexico population, a strong bottleneck after re-introduction, and/or rapid adaptation of a re-introduced population. Indications of a bottleneck (low genetic diversity or inbreeding) are not apparent (Table B.7). While southern Colorado and New Mexico populations are more genetically similar to each other than to the rest of Colorado (Fig. 3e), the distinctness of the New Mexico population suggests gene flow from southern Colorado populations alone would be unlikely to result in the pattern we observe here. We cannot determine whether the adaptive divergence observed in the New Mexico population represents rapid evolution of the re-introduced population, or introgression of some locally adapted historical New Mexico genetic variation into the re-introduced population. The potential for rapid adaptation would be a positive sign for a species facing a rapidly changing environment. However, the possibility of rapid evolution will require further investigation, potentially by evaluating other successful re-introductions (Braun et al. 2011) for signals of selection and measures of genetic diversity. Similarly, the possibility of ancestral locally adapted New Mexico genetic variation, may require examination of genetic samples from museum specimens or historical samples collected prior to the 1981 re-introduction, if those specimens exist (Braun and Williams 2015).
Predicted increased variability in weather under climate change scenarios could be especially challenging for alpine-obligate species likely to be locally adapted to harsh and highly seasonal conditions. The white-tailed ptarmigan will experience unique climate-related challenges due to a distribution that spans a broader latitude and elevation gradient than most other alpine specialists. Our findings suggest adaptive divergence among and within subspecies of white-tailed ptarmigan associated with vegetation, elevation, latitude, phenology, temperature, and precipitation. Our next steps include confirming selection is occurring relative to the identified environmental variables, many of which are highly correlated (Tables B.5 and B.6) and investigating more direct links between gene function and fitness (i.e., expression analyses, candidate gene investigation, etc.). Similarly, putative functional links (i.e., GO terms) are based on low density genome scans and linkage blocks (r2~0.10 at 100 kbp; Fig. B.3) that undoubtedly include additional potential targets of selection.
From a conservation perspective, our findings provide additional support for two of the subspecies (Vancouver and Southern) and potentially populations at the southern range extreme, as distinct units or potential sources of preadapted genetic variation that may facilitate potential range shifts, assisted migration, or translocation efforts. For example, the Southern and Vancouver subspecies have genetic variation associated with different precipitation and temperature regimes than the rest of the range. If local predictions of climate change indicate increased winter precipitation in an area, Vancouver may be a good genetic source. If summer temperatures are predicted to increase, populations within the Southern subspecies may be the most appropriate source population. Similarly, if there is genetic variation underlying molting phenology, the Southern subspecies may be a good source population for portions of the range that historically experience later snow melt or green-up, as was previously suggested for other species that undergo seasonal color change molts (Mills et al. 2018). Unfortunately, there is subdivision and local declines in the Southern subspecies signaling the potential loss of genetic variation for early phenology, high elevation, and low latitudes (based on clustering of individuals by our enriched functional group; Fig. 4c), that may otherwise be useful in assisted migration scenarios. Although the discontinuous distribution of alpine sky islands will likely impede a northward range shift, signals of selection in re-introduced populations (New Mexico) suggest the potential for rapid evolution and possibility of range shifts into previously unoccupied areas. The potential for outbreeding depression as a result of conservation actions remains given the suggestion of local adaptation for elevation and latitude. Parsing the potential for an evolutionary response to climate change in white-tailed ptarmigan, and possibly other alpine obligates, will require further study. Nevertheless, we provide a first look at environmentally associated adaptive divergence for an alpine obligate in relation to climate change, identify some physiological pathways for deeper investigation, and point to some conservation implications.
Data availability
All data were originally published in Langin et al. (2018). Molecular datasets were archived at https://doi.org/10.5066/f7gm86gz (Oyler-McCance et al. 2018) and genomic sequencing data were deposited in GenBank (biosample accession numbers: SAMN08132751-SAMN08132845).
References
Appella E, Weber IT, Blasi F (1988) Structure and function of epidermal growth factor-like regions in proteins. FEBS Lett 231:1–4
Berteaux D, Réale D, McAdam AG, Boutin S (2004) Keeping pace with fast climate change: can arctic life count on evolution? Integr Comp Biol 44:140–151
Bi K, Linderoth T, Singhal S, Vanderpool D, Patton JL, Nielsen R et al. (2019) Temporal genomic contrasts reveal rapid evolutionary responses in an alpine mammal during recent climate change. PLoS Genet 15:e1008119
Bivand R, Piras G (2015) Comparing implementations of estimation methods for spatial econometrics. J Stat Softw 63:1–36
Brauer CJ, Hammer MP, Beheregaray LB (2016) Riverscape genomics of a threatened fish across a hydroclimatically heterogeneous river basin. Mol Ecol 25:5093–5113
Braun CE, Hoffman RW, Rogers GE (1976) Wintering areas and winter ecology of white-tailed ptarmigan in Colorado, Colorado Division of Wildlife, Denver, CO, USA. Special Report 38
Braun CE, Taylor WP, Ebbert SE, Kaler RSA, Sandercock BK (2011) Protocols for successful translocation of ptarmigan. In: Watson RT, Cade TJ, Fuller M, Hunt G, Potapov E (eds) Gyrfalcons and ptarmigan in a changing world. The Peregrine Fund, Boise, ID, USA, p 339–348
Braun CE, Williams III SO (2015) History and status of the white-tailed ptarmigan in New Mexico. West Birds 46:233–243
Brown RD, Brasnett B (2010) Canadian Meteorological Centre (CMC) Daily Snow Depth Analysis Data, version 1. Colorado USA NASA National Snow and Ice Data Center, Distributed Active Archive Center, Boulder, CO, USA
Browning SR, Browning BL (2007) Rapid and accurate haplotype phasing and missing data inference for whole genome association studies by use of localized haplotype clustering. Am J Hum Genet 81:1084–1097
Bryant JP, Kuropat PJ (1980) Selection of winter forage by subarctic browsing vertebrates: the role of plant chemistry. Annu Rev Ecol Syst 11:261–285
Capblancq T, Luu K, Blum MGB, Bazin E (2018a) How to make use of ordination methods to identify local adaptation: a comparison of genome scans based on PCA and RDA. bioRxiv: 258988v2
Capblancq T, Luu K, Blum MGB, Bazin E (2018b) Evaluation of redundancy analysis to identify signatures of local adaptation. Mol Ecol Resour 18:1223–1233
Carey C, Martin K (1997) Physiological ecology of incubation of ptarmigan eggs at high and low altitudes. Wildlife Biol 3:211–218
Charmantier A, McCleery RH, Cole LR, Perrins C, Kruuk LEB, Sheldon BC (2008) Adaptive phenotypic plasticity in response to climate change in a wild bird population. Science 320:800–803
Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L et al. (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6:80–92
Clarke JA, Johnson RE (1992) The influence of spring snow depth on white-tailed ptarmigan breeding success in the Sierra Nevada. Condor 94:622–627
Coop G, Witonsky D, Di Rienzo A, Pritchard JK (2010) Using environmental correlations to identify loci underlying local adaptation. Genetics 185:1411–23
Dalloul RA, Long JA, Zimin AV, Aslam L, Beal K, Blomberg LA et al. (2010) Multi-platform next-generation sequencing of the domestic Turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biol 8:e1000475
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA et al. (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158
Dragon S, Carey C, Martin K, Baumann R (1999) Effect of high altitude and in vivo adenosine/beta-adrenergic receptor blockade on ATP and 2,3BPG concentrations in red blood cells of avian embryos. J Exp Biol 202:2787–2795
Dray S, Blanchet D, Borcard D, Clappe S, Guenard G, Jombart T et al. (2019) Adespatial: multivariate multiscale spatial analysis. R package version 0.3-4. http://cran.r-project.org/package=adespatial
Erikstad KE, Andersen R (1983) The effect of weather on survival, growth rate, and feeding time in different sized willow grouse broods. Ornis Scand 14:249–252
Fabian DK, Kapun M, Nolte V, Kofler R, Schmidt PS, Schlötterer C et al. (2012) Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Mol Ecol 21:4748–4769
Fedy BC, Martin K (2011) The influence of fine-scale habitat features on regional variation in population performance of alpine white-tailed ptarmigan. Condor 113:306–315
Fedy BC, Martin K, Ritland C, Young J (2008) Genetic and ecological data provide incongruent interpretations of population structure and dispersal in naturally subdivided populations of white-tailed ptarmigan (Lagopus leucura). Mol Ecol 17:1905–1917
Fick SE, Hijmans RJ (2017) Worldclim 2: new 1-km spatial resolution climate surfaces for global land areas. Int J Climatol 37:4302–4315
Forester BR, Lasky JR, Wagner HH, Urban DL (2018) Comparing methods for detecting multilocus adaptation with multivariate genotype-environment associations. Mol Ecol 27:2215–2233
Forester BR, Jones MR, Joost S, Landguth EL, Lasky JR (2016) Detecting spatial genetic signatures of local adaptation in heterogenous landscapes. Mol Ecol 24:104–120
Frederick GP, Gutiérrez RJ (1992) Habitat use and population characteristics of the white-tailed ptarmigan in the Sierra Nevada, California. Condor 94:889–902
Freeman BG, Lee-Yaw JA, Sunday JM, Hargreaves AL (2018) Expanding, shifting and shrinking: the impact of global warming on species’ elevational distributions. Glob Ecol Biogeogr 27:1268–1276
Friedl MA, Gray J, Sulla-Menashe D (2009) MCD12Q2 MODIS/Terra+Aqua Land Cover Dynamics Yearly L3 Global 500m SIN Grid V005 [2000–2010]. NASA EOSDIS Land Processes DAAC. Sioux Falls, SD, USA
García-González R, Aldezabal A, Laskurain NA, Margalida A, Novoa C (2016) Influence of snowmelt timing on the diet quality of pyrenean rock ptarmigan (Lagopus muta pyrenaica): implications for reproductive success. PLoS ONE 11:1–21
Giesen KM, Braun CE, May TA (1980) Reproduction and nest-site selection by white-tailed ptarmigan in Colorado. Wilson Bull 92:188–199
Hakkarainen H, Virtanen R, Honkanen JO, Roininen H (2007) Willow bud and shoot foraging by ptarmigan in relation to snow level in NW Finnish Lapland. Polar Biol 30:619–624
Hall D, Salomonson V, Riggs G (2006) MODIS/Terra Snow Cover Daily L3 Global 500m Grid, Version 5. NASA Natl Snow Ice Data Cent Distrib Act Arch Center Boulder, CO, USA
Hannon SJ, Eason PK, Martin K (1998) Willow ptarmigan (Lagopus lagopus), version 2.0. In: Poole AF, Gill FB (eds) The Birds of North America. Cornell Lab of Ornithology, Ithaca, New York, NY, USA
Henry P, Sim Z, Russello MA (2012) Genetic evidence for restricted dispersal along continuous altitudinal gradients in a climate change-sensitive mammal: The American Pika. PLoS ONE 7:1–10
Hoffman RW, Braun CE (1975) Migration of a wintering population of white-tailed ptarmigan in Colorado. J Wildl Manage 39:485–490
Hoffmann AA, Sgró CM (2011) Climate change and evolutionary adaptation. Nature 470:479–485
Holsinger LM, Parks SA, Parisien M-A, Miller C, Batllori E, Moritz MA (2019) Climate change likely to reshape vegetation in North America’s largest protected areas. Conserv Sci Pract 1:e50
Höst P (1942) Effect of light on the moults and sequences of plumage in the willow ptarmigan. Auk 59:388–403
Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D et al. (2007) DAVID bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 35:169–175
Imperio S, Bionda R, Viterbi R, Provenzale A (2013) Climate change and human disturbance can lead to local extinction of alpine rock ptarmigan: new insight from the Western Italian Alps. PLoS ONE 8:e81598
Jacobsen EE, White CM, Emison WB (2007) Molting adaptations of rock ptarmigan on Amchitka Island, Alaska. Condor 85:420
Kawecki T, Ebert D (2004) Conceptual issues in local adaptation. Ecology Letters 7:1225–1241
Ke J, Wang L, Xiao D (2017) Cardiovascular adaptation to high-altitude hypoxia. In: Zheng J, Zhou C (eds) Hypoxia and human diseases. IntechOpen Limited, London, UK, p 117–134
Klanderud K, Birks HJB (2003) Recent increases in species richness and shifts in altitudinal distributions of Norwegian mountain plants. Holocene 13:1–6
Kohl KD, Varner J, Wilkening JL, Dearing MD (2018) Gut microbial communities of American pikas (Ochotona princeps): evidence for phylosymbiosis and adaptations to novel diets. J Anim Ecol 87:323–330
Kozma R, Rödin-Mörch P, Höglund J (2019) Genomic regions of speciation and adaptation among three species of grouse. Sci Rep 9:1–8
Laiolo P, Obeso JR (2017) Life-history responses to the altitudinal gradient. In: Catalan J (ed) High mountain conservation in a changing world, Advances in Global Change Research, Vol 62, p. 3–36. Springer, Cham
Langin KM, Aldridge CL, Fike JA, Cornman RS, Martin K, Wann GT et al. (2018) Characterizing range-wide divergence in an alpine-endemic bird: a comparison of genetic and genomic approaches. Conserv Genet 19:1471–1485
Latifovic R, Pouliot D, Olthof I (2017) Circa 2010 land cover of Canada: local optimization methodology and product development. Remote Sens 9:1098
Legendre P, Legendre L (2012) Numerical Ecology, 3rd edn. Elsevier, Amsterdam, The Netherlands
Mangin B, Siberchicot A, Nicolas S, Doligez A, This P, Cierco-Ayrolles C (2012) Novel measures of linkage disequilibrium that correct the bias due to population structure and relatedness. Heredity 108:285–291
Martin K, Brown GA, Young JR (2004) The historic and current distribution of the Vancouver Island white-tailed ptarmigan (Lagopus leucurus saxatilis). J F Ornithol 75:239–256
Martin K, Robb LA, Wilson S, Braun CE (2015) White-tailed ptarmigan (Lagopus leucura), version 2.0. In: Rodewald PG (ed) The Birds of North America. Cornell Lab of Ornithology, Ithaca, New York, NY, USA
Martin K, Wiebe KL (2004) Coping mechanisms of alpine and arctic breeding birds: extreme weather and limitations to reproductive resilience. Integr Comp Biol 44:177–185
May TA, Braun CE (1972) Seasonal foods of adult white-tailed ptarmigan in Colorado. J Wildl Manage 36:1180–1186
McKinnon L, Picotin M, Bolduc E, Juillet C, Bêty J (2012) Timing of breeding, peak food availability, and effects of mismatch on chick growth in birds nesting in the High Arctic. Can J Zool 90:961–971
Mills LS, Bragina EV, Kumar AV, Zimova M, Lafferty DJR, Feltner J et al. (2018) Winter color polymorphisms identify global hot spots for evolutionary rescue from climate change. Science 359:1033–1036
Montgomerie R, Holder K (2008) Rock ptarmigan (Lagopus muta), version 2.0. In: Poole AF (ed) The Birds of North America. Cornell Lab of Ornithology, Ithaca, New York, NY, USA
Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, Schäffer AA (2008) Database indexing for production MegaBLAST searches. Bioinformatics 24:1757–1764
Moss R (1973) The digestion and intake of winter foods by wild ptarmigan in Alaska. Condor 75:293–300
Moss R (1974) Winter diets, gut lengths, and interspecific competition in Alaskan ptarmigan. Auk 91:737–746
Moss R (1983) Gut size, body weight, and digestion of winter foods by grouse and ptarmigan. Condor 85:185–193
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York, NY, USA
New Mexico Department of Game and Fish (2016) White-tailed ptarmigan (Lagopus leucura) recovery plan. New Mexico Department of Game and Fish, Wildlife Management Division, Santa Fe, NM, USA
NOAA National Centers for Environmental Prediction (NCEP) (2018) NCEP-NCAR Reanalysis montly zonal and meridional winds at standard pressure levels on a 2.5 lat/lon grid. NOAA National Centers for Environmental Prediction (NCEP), College Park, MD, USA
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D et al. (2017) vegan: community ecology package version 2.4-3. https://cran.r-project.org/package=vegan
Oyler-McCance SJ, Langin KM, Cornman RS, Fike J, Aldridge CL, Martin KM et al. (2018) Sample collection information, single nucleotide polymorphism, and microsatellite data for white-tailed ptarmigan across the species range generated in the Molecular Ecology Lab during 2016: U.S. Geological Survey data release, https://doi.org/10.5066/F7GM86GZ
Palo RT (1984) Distribution of birch (Betula spp.), willow (Salix spp.), and poplar (Populus spp.) secondary metabolites and their potential role as chemical defense against herbivores. J Chem Ecol 10:499–520
Paradis E, Schlier K (2018) ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35:526–528
Parmesan C, Yohe G (2003) A globally coherent fingerprint of climate change. Nature 421:37–42
Pedersen S, Odden M, Pedersen HC (2017) Climate change induced molting mismatch? Mountain hare abundance reduced by duration of snow cover and predator abundance. Ecosphere 8:e01722
Pepin N, Bradley RS, Diaz HF, Baraer M, Caceres EB, Forsythe N et al. (2015) Elevation-dependent warming in mountain regions of the world. Nat Clim Chang 5:424–430
Persons NW, Hosner PA, Meiklejohn KA, Braun EL, Kimball RT (2016) Sorting out relationships among the grouse and ptarmigan using intron, mitochondrial, and ultra-conserved element sequences. Mol Phylogenet Evol 98:123–132
Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE (2012) Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7:e37135
Pörtner HO (2002) Climate variations and the physiological basis of temperature dependent biogeography: systemic to molecular hierarchy of thermal tolerance in animals. Comp Biochem Physiol part A 132:739–761
Price N, Lopez L, Platts AE, Lasky JR (2020) In the presence of population structure: from genomics to candidate genes underlying local adaptation. Ecol Evol 10:1889–1904
Pyle P (2007) Revision of molt and plumage terminology in ptarmigan (Phasianidae: Lagopus spp.) based on evolutionary considerations. Auk 124:508
Rellstab C, Gugerli F, Eckert AJ, Hancock AM, Holderegger R (2015) A practical guide to environmental association analysis in landscape genomics. Mol Ecol 24:4348–4370
Resano-Mayor J, Korner-Nievergelt F, Vignali S, Horrenberger N, Barras AG, Braunisch V et al. (2019) Snow cover phenology is the main driver of foraging habitat selection for a high-alpine passerine during breeding: implications for species persistence in the face of climate change. Biodivers Conserv 28:2669
Rolando A, Laiolo P, Formica M (1997) A comparative analysis of the foraging behaviour of the chough Pyrrhocorax pyrrhocorax and the Alpine chough Pyrrhocorax graculus coexisting in the Alps. Ibis 139:461–467
Rundel PW, Millar CI (2016) Alpine Ecosystems. In: Zavaleta E, Mooney H (eds) Ecosystems of California. University of California, Berkeley, CA, USA, p 613–634
Salomonsen F (1936) On a new race of willow grouse. Bull Br Ornithol Club 56:99–100
Singh CP (2008) Alpine ecosystems in relation to climate change. ISG Newsl 14:54–55
Slatkin M (2008) Linkage disequilibrium: understanding the genetic past and mapping the medical future. Nat Rev Genet 9:477–485
Somero G (2010) The physiology of climate change: how potentials for acclimatization and genetic adaptation will determine ‘winners’ and ‘losers. J Exp Biol 213:912–920
Spear SL, Aldridge CL, Wann GT, Braun CE (2019) Fine-scale habitat selection by breeding white-tailed ptarmigan in Colorado. J Wildl Manage 84:172–184
Stokken K-A (1993) Energetics and adaptations to cold in ptarmigan in winter. Ornis Scand 23:366–370
Storz JF (2005) Using genome scans of DNA polymorphism to infer adaptive population divergence. Mol Ecol 14:671–688
Swanson DL, King MO, Harmon E (2014) Seasonal variation in pectoralis muscle and heart myostatin and tolloid-like proteinases in small birds: a regulatory role for seasonal phenotypic flexibility? J Comp Physiol B 184:249–258
Thornton PE, Running SW, White MA (1997) Generating surfaces of daily meteorological variables over large regions of complex terrain. J Hydrol 190:214–251
Thornton PE, Thornton MM, Mayer BW, Wei Y, Devarakonda R, Vose RS et al. (2018) Daymet: daily surface weather data on a 1-km grid for North America, version 3. Oak Ridge, Tennessee, USA
Thuiller W (2004) Patterns and uncertainties of species’ range shifts under climate change. Glob Change Biol 10:2020–2027
United States Fish and Wildlife Service (2012) Endangered and threatened wildlife and plants; 90-day finding on a petition to list the southern white-tailed ptarmigan and the Mt. Rainier white-tailed ptarmigan as threatened with critical habitat. Fed Regist 77:33143–33155
United States Geological Survey EROS Center (2007) North American elevation 1-kilometer resolution, 3rd edn. National Atlas of the US, Reston, VA
Vos PG, Paulo MJ, Voorrips RE, Visser RGF, van Eck HJ, van Eeuwijk FA (2017) Evaluation of LD decay and various LD‑decay estimators in simulated and SNP‑array data of tetraploid potato. Theor Appl Genet 130:123–135
Walker WP, Aradhya S, Hu C-L, Shen S, Zhang W, Azarani A et al. (2007) Genetic analysis of attractin homologs. Genesis 45:744–756
Walker WP, Gunn TM (2010) Shades of meaning: the pigment-type switching system as a tool for discovery. Pigment Cell Melanoma Res 23:485–495
Wang G, Hobbs NT, Galbraith H, Giesen KM (2002a) Signatures of large-scale and local climates on the demography of white-tailed ptarmigan in Rocky Mountain National Park, Colorado, USA. Int J Biometeorol 46:197–201
Wang G, Hobbs NT, Giesen KM, Galbraith H, Ojima DS, Braun CE (2002b) Relationships between climate and population dynamics of white-tailed ptarmigan Lagopus leucurus in Rocky Mountain National Park, Colorado, USA. Clim Res 23:81–87
Wann GT, Aldridge CL, Braun CE (2014) Estimates of annual survival, growth, and recruitment of a white-tailed ptarmigan population in Colorado over 43 years. Popul Ecol 56:555–567
Wann GT, Aldridge CL, Braun CE (2016) Effects of seasonal weather on breeding phenology and reproductive success of alpine ptarmigan in Colorado. PLoS ONE 11:e0158913
Wann GT, Aldridge CL, Seglund AE, Oyler‐McCance SJ, Kondratieff BC, Braun CE (2019) Mismatches between breeding phenology and resource abundance of resident alpine ptarmigan negatively affect chick survival. Ecol Evol 9:7200–7212
Weeden RB (1967) Seasonal and geographic variation in the foods of adult white-tailed ptarmigan. Condor 69:303–309
Werhahn G, Liu Y, Meng Y, Cheng C, Lu Z, Atzeni L et al. (2020) Himalayan wolf distribution and admixture based on multiple genetic markers J Biogeogr https://doi.org/10.1111/jbi.13824
Wiebe KL, Martin K (1998) Costs and benefits of nest cover for ptarmigan: changes within and between years. Anim Behav 56:1137–1144
Wilson S, Martin K (2008) Breeding habitat selection of sympatric white-tailed, rock and willow ptarmigan in the southern Yukon Territory, Canada. J Ornithol 149:629–637
Wilson S, Martin K (2011) Life-history and demographic variation in an alpine specialist at the latitudinal extremes of the range. Popul Ecol 53:459–471
Xin J-W, Chai Z-X, Zhang C-F, Zhang Q, Zhu Y, Cao H-W et al. (2019) Transcriptome profiles revealed the mechanisms underlying the adaptation of yak to high-altitude environments. Sci Rep 9:7558
Zimova M, Hackländer K, Good JM, Melo-Ferreira J, Alves PC, Mills LS (2018) Function and underlying mechanisms of seasonal colour moulting in mammals and birds: what keeps them changing in a warming world? Biol Rev 93:1478–1498
Acknowledgements
We are grateful for the people and institutions that collected and shared samples from across the white-tailed ptarmigan range for the original Langin et al. (2018) study: Jennifer A. Fike, Kathy Martin, Amy E. Seglund, Michael A. Schroeder, Clait E. Braun, David P. Benson, Brad C. Fedy, Jessica R. Young, Scott Wilson, Donald H. Wolfe, Kathryn Bernier, Sharon Birks, John Bulger, Ray Collingwood, Avery Cook, Sarah Hudson, Doug Jury, Lee Kaiser, Richard Merizon, Jason Robinson, Serena Rocksund, William Taylor, the University of Washington Burke Museum, Colorado Parks and Wildlife, Utah Division of Wildlife Resources, and Alaska Department of Fish and Game. This work was supported by the U.S. Fish and Wildlife Service, U.S. Geological Survey, and Colorado State University. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. government.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Xiangjiang Zhan
Supplementary information
Rights and permissions
About this article
Cite this article
Zimmerman, S.J., Aldridge, C.L., Langin, K.M. et al. Environmental gradients of selection for an alpine-obligate bird, the white-tailed ptarmigan (Lagopus leucura). Heredity 126, 117–131 (2021). https://doi.org/10.1038/s41437-020-0352-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-020-0352-6
This article is cited by
-
Conservation genomics of an endangered arboreal mammal following the 2019–2020 Australian megafire
Scientific Reports (2023)