Abstract
Reaction norms or tolerance curves have often been used to predict how organisms deal with fluctuating environments. A potential drawback is that reaction norms measured in different constant environments may not capture all aspects of organismal responses to fluctuating environments. We examined growth of the filamentous fungus Neurospora crassa in fluctuating temperatures and tested if growth in fluctuating temperatures can be explained simply by the growth in different constant temperatures or if more complex models are needed. In addition, as previous studies on fluctuating environments have revealed that past temperatures that organisms have experienced can affect their response to current temperature, we tested the roles of different epigenetic mechanisms in response to fluctuating environments using different mutants. We found that growth of Neurospora can be predicted in fluctuating temperatures to some extent if acclimation times are taken into account in the model. Interestingly, while fluctuating environments have been linked with epigenetic responses, we found only some evidence of involvement of epigenetic mechanisms on tolerating fluctuating temperatures. Mutants which lacked H3K4 or H3K36 methylation had slightly impaired response to temperature fluctuations, in addition the H3K4 methylation mutant and a mutant in the RNA interference pathway had altered acclimation times.
Similar content being viewed by others
Introduction
Phenotypic plasticity is the ability of organisms to change or develop different phenotypes in response to environmental changes (DeWitt and Scheiner 2004). We can describe phenotypic plasticity using reaction norms, where the phenotypic value of a particular trait is a function of an environmental variable. For example, the growth of a single genotype in different temperatures. Natural selection can act on plasticity (Scheiner 2002; Nussey et al. 2005) and plasticity is important in life history theory (Day and Rowe 2002) and sexual selection theory (Ingleby et al. 2010). Plasticity can also influence evolutionary dynamics when a population adapts to a new environment (Lande 2009; Chevin et al. 2010). In terms of practical applications, reaction norms for temperature, also called thermal performance curves, have been used in predicting the effects of climate change on extinction risk in ectotherm populations (Deutsch et al. 2008; Kingsolver et al. 2013; Vasseur et al. 2014; Levy et al. 2015).
Reaction norms are a useful tool, but there exists a significant complication to their practical application. Reaction norms are often measured in the laboratory in constant environmental conditions. The wild most environments rarely stay constant for long, day and night cycles, seasons and daily changes in weather all cause the environment to change constantly. This may be a significant drawback for the usefulness of reaction norms, as recently there have been reports that reaction norms measured in constant environments do not predict the performance of organisms in fluctuating environments (Kingsolver et al. 2009, 2015; Niehaus et al. 2012; Rezende et al. 2014; Ketola et al. 2014; Ketola and Saarinen 2015 and Saarinen et al. 2018). Ketola and Kristensen (2017) and Sinclair et al. (2016) list the key complications of why reaction norms measured in the lab may not predict the performance in natural environments; performance depends on the current temperature, but also on temperatures experienced in the past (Schulte et al. 2011; Rezende et al. 2014); the time an organism has experienced the current temperature may influence acclimation, as acclimation to a given temperature may not be instantaneous (Johnston and Dunn 1987). The duration of exposure or the frequency of environmental fluctuations may also be important (Ketola and Kristensen 2017). For example, the benefits of inducible plasticity could be diminished if the response time for phenotypic adjustment is slower than a new alteration in the environment (Padilla and Adolph 1996).
Mechanisms of thermal tolerance have been studied intensively, and in general, heat shock proteins are involved in tolerance responses in many organisms (Feder and Hofmann 1999). In fungi, thermal tolerance is mediated by heat shock proteins (Piper 1993; Glatz et al. 2015), other metabolites, in particular the sugar trehalose (De Virgilio et al. 1994; Bonini et al. 1995; Arguelles 1997) and changes in lipid bilayer composition (Glatz et al. 2015). However, the regulation of thermotolerance is not completely understood. Epigenetic mechanisms are one possibility for indirectly mediating the influence of past environments. These include chromatin modifications such as DNA methylation (Bird 2002), and different types of histone modifications (Bannister and Kouzarides 2011). It is known that epigenetic mechanisms can mediate cell memory (Bird 2002; Gaydos et al. 2014; de la Paz Sanchez et al. 2015) and plasticity (Kooke et al. 2015; Kronholm et al. 2016), and are required for certain parental effects (Wibowo et al. 2016) or between generation plasticity (Luna and Ton 2012; Rasmann et al. 2012). Effects that are passed from one generation to the next can create lag in fluctuating environments. However, memory effects can also occur within one generation. If epigenetic mechanisms control phenotypic plasticity, they may be needed in fluctuating environments. In particular, we have previously observed that the set-2 mutant, which is deficient in histone 3 lysine 36 methylation, and the qde-2 mutant, which is defective in milRNA processing, are both impaired in their temperature responses in constant temperatures (Kronholm et al. 2016).
This study has two aims: the first is to investigate if reaction norms measured in constant temperatures can be used to explain the growth rate of the filamentous fungus Neurospora crassa in fluctuating temperatures, and if not, why do reaction norms measured in constant environments fail? The second is to investigate the role of epigenetic mechanisms within a generation in fluctuating environments: does the absence of certain chromatin modifications change the shape of reaction norms in fluctuating environments, indicating that this specific mechanism is needed in a fluctuating environment? In order to investigate the importance of epigenetic mechanisms in response to fluctuating temperatures, we used a collection of deletion mutants that lacked a certain epigenetic mechanism and grew these mutants in the different fluctuating environments. The hypothesis is that if an epigenetic mechanism is involved in sensing temperature change or in the expression of acclimation, mutants deficient in this mechanism would acclimate slower or ineffectively. If an epigenetic mechanism is involved in memory of past environments and causes lag in acclimation to a new environment, then mutants deficient in this mechanism would acclimate faster.
As a fluctuating environment, we used two temperatures 30 and 40 °C and durations ranging from 30 to 720 min the cultures spent in each temperature. The species N. crassa occurs mainly on burned vegetation in tropical and sub-tropical regions (Turner et al. 2001), and its optimum growth temperature is around 35 °C, while 40 °C is a stressful temperature for it (Kronholm et al. 2016). We also investigated how long it takes for the fungus to acclimate from one temperature to another, that is, to reach a constant growth rate in the new temperature. We then tested different growth models to predict growth in fluctuating environments, such as models with no lag in acclimation or lag effects included.
We find that reaction norms measured in constant environments predict growth rate poorly in rapidly fluctuating environments, due to time lag in temperature acclimation. Better predictions are obtained from a model that takes lag times into account. Epigenetic mechanisms, in particular histone modifications H3K36 methylation and H3K4 trimethylation, affect growth in fluctuating environments, but are not as important as in constant environments. We also found that H3K4 trimethylation and RNA interference pathway are involved in acclimation responses.
Materials and methods
Neurospora strains and growth measurements
We previously obtained a set of deletion mutants from the Neurospora knockout mutant collection (Colot et al. 2006) from Fungal Genetics Stock Center (FGSC) (McCluskey et al. 2010), the qde-2 mutant was provided by Tereza Ormsby and Eric Selker, and backcrossed these mutants to the wild-type strain 2489 (Kronholm et al. 2016). Backcrossing was done so that mutants differed from the control genotype only in the deleted gene. Mutants used in this study are listed in Table 1; all strains were of mating type A. Procedures used for backcrossing and confirming the presence of the deletions by PCR can be found in Kronholm et al. (2016). The experiment contained 20 mutants and the wild-type strain 2489, 21 strains in total.
We grew the strains on Vogel’s standard growth medium (Metzenberg 2003) with 1.5% agar in disposable 25-ml pipettes prepared by the method of White and Woodward (1995). At the start of each experiment, a strain was inoculated at the other end of the pipette and the growth of a strain was followed by marking the position of the mycelial front at a given time point. Growth rate was obtained as the slope of the regression line for time against distance the mycelium had grown. Detailed description of the growth measurements can be found in Kronholm et al. (2016).
Growth in fluctuating environments
To measure growth rate in fluctuating environments, we used programmable growth chambers (MTM-313 Plant Growth Chamber, HiPoint Corp., Taiwan) to change the temperature periodically. We used fluctuations with an amplitude of 10 °C from 30 to 40 °C. We used eight different fluctuation regimes; cultures spent either 30, 60, 90, 120, 150, 240, 480, or 720 min in one temperature during one cycle. Thus fluctuations had periods of 1, 2, 3, 4, 5, 8, 16, or 24 h. Throughout this study, when discussing the different fluctuation regimes, we will refer to the time cultures spent in one temperature as duration of a step. Growth was followed for a period of 104 h and measurements were taken every 8 and 16 h to obtain growth rates for different step durations (Supplementary figure S1A). In each fluctuation regime, the temperature changed in a stepwise manner (Supplementary figure S2), so that during one cycle cultures spent half of the time in 30 °C and half of time in 40 °C. Thus the mean temperature for each treatment was 35 °C and the total time spent in 30 and 40 °C is the same.
We grew 21 strains in the eight different fluctuation regimes with five replicates each; the strain nst-7 was missing from step durations 240, 480, and 720 min, giving 825 growth assays in total. With two growth chambers with three independently controlled compartments each, we had six different compartments available. To control for compartment effects, we rotated the fluctuation regimes among the compartments between different replicates. Strains were always randomised within a compartment.
In a preliminary experiment, we measured the rate of temperature equilibration between growth chamber air and the agar tubes used to measure growth rates of the strains. We inserted a temperature probe into the agar of a growth tube and monitored the temperature in the growth tube and the air temperature in the growth chamber during heating and cooling. It took about 12 min for the growth chamber air temperature to rise from 30 °C to nearly 40 °C, while the same temperature change took ∼20 min for the agar (Supplementary figure S3). During heating, the growth chamber first increased the temperature nearly to the set point, but to prevent overshooting, increasing the temperature to the last remaining degree took most of the elapsed time. Cooling was more efficient and it took ∼6 min for the agar temperature to drop from 40 to 30 °C. Therefore, the actual times the cultures spent in 40 and 30 °C were shorter than the nominal temperature settings, and cycles of 30 min at each temperature were the shortest fluctuations possible that could be achieved with our system.
Temperature shift experiments
We performed temperature shift experiments to measure how the growth rate changed, and if there was a lag time when Neurospora acclimated to a new temperature. In a shift experiment, we initially inoculated the cultures and grew them in either constant 30 or 40 °C for 24 h to allow the cultures to acclimate to the current temperature. Then we swapped the cultures between the two different incubators, those cultures that had been initially growing in 30 °C were moved to 40 °C and vice versa. As the growth chambers were constantly at the same temperature and the cultures were shifted, there was no time lag associated with heating or cooling the growth chamber, only the heat transferred to agar medium itself. After the shift, we recorded the growth of the cultures every hour for 8 h and for two additional time points, 25 and 32 h, to allow fine grain monitoring of changes in growth rate after the shift (Supplementary figure S1B). For analysis we calculated growth rates for 2 h intervals, as recording 1 hour of growth in the tubes was difficult as the marks were very close to each other and thus the data measured every 2 h provided a reasonable smoothing of measurement noise.
We performed two shift experiments. In the first experiment, we used two genotypes: the wild-type control genotype 2489 and the qde-2 mutant, both genotypes were replicated 10 times in both shifts, giving 40 growth assays in total. We included qde-2 in this initial experiment, as our previous results suggested that it is involved in temperature responses (Kronholm et al. 2016). In the second experiment, we performed a temperature shift experiment with all of the mutants except nst-7, dcl-1, and dcl-2. The dcl single mutants were excluded because in all previous experiments they had the same phenotype as the dcl-1 dcl-2 double mutant. In the second experiment, each of the 18 genotypes was replicated 4 times in both shifts, giving 144 growth assays in total.
Estimating the cell cycle durations
Neurospora is a filamentous organism, its cells remain cytoplasmically connected to each other and the nuclei can move within the mycelium. Thus, asexual mitotic divisions do not constitute generations as they do in unicellular microbes, and estimating the number of mitotic divisions that occurred for a certain amount of growth is not simple. In addition, mitotic divisions are not fully synchronous, and it is not known how many of the nuclei are actively dividing in a growing hyphae. The duration of cell cycle has been estimated to be 103 min (Martegani et al. 1981) in conditions that correspond to a growth rate of 4.4 mm/h (Ryan et al. 1943; Kronholm et al. 2016) and around 217 min (Martegani et al. 1981) in conditions where growth rate is around 2.3 mm/h (Ryan et al. 1943). Based on these numbers we interpolated cell cycle durations for our observed growth rates assuming a linear relationship. These numbers should be regarded as rough estimates that are only meant to give an approximate idea of what is the relationship between step duration and cell cycle duration.
Data analysis
Analysis of growth rates in fluctuating environments
We used a linear mixed model to investigate the effects of the different fluctuation regimes on the growth of the different strains. We first fitted the growth chamber compartment effect to the data and removed the compartment effect by subtracting the effect from the raw data. Because the strains and treatments were randomly distributed, removing the average effect of compartment does not remove the strain or treatment effects. We encoded the different step durations as factors to allow us to fit the same model for all strains, as responses were non-linear. The model was
where μ is the intercept, Fi is the ith step duration, Gj is the jth genotype, \(\epsilon\) is the residual, and yij is the growth rate of the jth genotype in step duration i. The duration of the step was fitted as a fixed effect and genotype as a random effect. The mixed model was fitted with the “lmer” function in R (R Core Team 2013) and statistical testing was performed with the “lmerTest” package (Kuznetsova et al. 2015). The lmerTest package implements F tests using type III sums of squares with Satterthwaite correction for degrees of freedom. For comparing the reaction norm shape for each mutant to the control, we used a pairwise ANOVA to test if the genotype by step duration interaction was significant. Correction for multiple testing was done using the Bonferroni–Holm method (Holm 1979).
Estimating lag times from temperature shift experiments
To obtain lag times from the temperature shift experiments, we fitted non-linear regression curves to the temperature shift data for each direction of temperature change using the model
where y is the growth rate, β0 is the asymptote or the final growth rate after acclimation, β1 is the amount of growth rate to be gained (i.e. β0 − β1 is the growth rate immediately after temperature change), β2 is the time it takes for growth rate to increase to half of its maximum value, t is the time elapsed since temperature change and \(\epsilon\) is the residual (Venables and Ripley 2002). This yielded a set of parameters β0, β1 and β2 that estimated for each direction of temperature change. Fitting a negative exponential function has the advantage that all the parameters have a biological interpretation. We are interested in the β2 parameter as it gives us a measure for the length of the lag time. When fitting the model to the shift from 30 to 40 °C data, we excluded data points from constant 30 °C and assumed that growth rate drops faster than the resolution in our data collection. For fitting the model and testing for differences in lag times among the different genotypes we used the “nls” and “gnls” functions in R.
Growth modelling based on temperature shift experiments
To model growth in fluctuating environments for the control genotype, 2489, we used the acclimation times obtained from temperature shift experiments and tested if it could predict growth in fluctuating environments with this data. After obtaining numerical estimates for Eq. (2) for both acclimation directions, we defined two lag functions giving the instantaneous growth rate as a function of time t elapsed since temperature change as
where i = 1 for a change from 30 to 40 °C and i = 2 for a change from 40 to 30 °C. Then we used growth rate models to predict growth in fluctuating environments. In our initial model, growth rate rp in a fluctuating environment where step duration was p hours during one cycle was
where the numerator gives the amount of growth (in mm) that occurs during one temperature cycle, l1(t) and l2(t) are the lag functions obtained from fitting Eq. (2) to the different temperature shifts and using the obtained β parameters. We integrate over the time spent in given temperature to obtain the predicted growth that occurs after the temperature shift, the denominator gives the total time (in h) for one full temperature cycle to obtain the growth rate as mm/h.
As our initial lag model worked poorly for fast fluctuations (see Results), we later refined our initial model to account for partial acclimation. The predicted lag times were much longer than the step durations in fast fluctuations. Therefore, acclimation happens only partially before the temperature changes again and consequently next acclimation does not begin from the growth rate the culture would eventually reach in constant temperature. We modified the model so that we updated the parameter β1, setting the amount of growth rate to be gained in the next cycle to reflect the growth rate reached in the previous temperature.
The refined model followed the following algorithm: a culture spent p hours first in 30 °C and we calculated the distance grown, then we calculated the current growth rate, r*, at time p. If r* < β0 of the next lag function (β0,2 during first cycle), then we set \(\beta _{1,2}^ \ast = \beta _{0,2} - r^ \ast\), otherwise we set \(\beta _{1,2}^ \ast = \beta _{1,2}\). We then calculated the distance grown using the updated l2 function. After p hours of growth in the next temperature, current growth rate was again calculated and it was checked if r* < β0,1, then we set \(\beta _{1,1}^ \ast = \beta _{0,1} - r^ \ast\) and \(\beta _{1,1}^ \ast = \beta _{1,1}\) otherwise. We then calculated the distance grown using the updated l1 function. The empirical measurements for the different strains in the fluctuating environments were done over 103 h, so the number of temperature cycles in the model, n, was determined by how many full cycles of 2p could fit into 103 h. The growth rate was obtained from
where the β1,i parameters were updated for each step k as described above.
We compared how well the simulation models fit the observed data by calculating mean squared deviations (MSD) from the observed data for each model. We can use MSD for comparing the relative ranks of the different models, but MSD does not tell us how well the model fits the data in an absolute sense. We chose to use MSD because traditional measures of model fit, like R2 or AIC, cannot be calculated for our simulated models.
Results
Effect of temperature fluctuations on growth
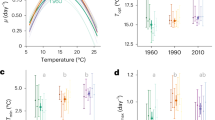
At constant 30 °C, the control genotype 2489 grows at 4.4 mm/h and at constant 40 °C at 2.4 mm/h (Kronholm et al. 2016). The control strain has its optimum at 35 °C (Fig. 1a). We observed that the growth rate of the control strain in step durations from 30 to 150 min was mainly determined by its growth rate in constant 40 °C (Fig. 1b), as growth rate of 2489 in these step durations was only slightly above 2.4 mm/h. Growth rate increased slightly with the period of fluctuations. Under the assumption that acclimation to 30 and 40 °C would be instantaneous and growth rate would change immediately to corresponding growth rate at constant temperature, the growth rate per hour in fluctuating temperature would be the average of the growth rates in constant 30 and 40 °C because under all fluctuation regimes, the cultures spend equal amount of time in 30 and 40 °C. This naive expected growth rate would be 3.38 mm/h, the observed growth rates in these fast fluctuations were well below this rate (Fig. 1b). When step durations increased to 240 min and above growth rate increased more rapidly, and at step durations 480 and 720 min, growth rate was very close to the expected growth rate of 3.38 mm/h.
a Reaction norm for the genotype 2489 in constant temperatures, n = 12 for each point. Data from Kronholm et al. (2016). b Reaction norm for the genotype 2489 in different step durations when temperature fluctuates between 30 and 40 °C, n = 5 for each point. Horizontal blue and red lines show growth rate at constant 30 and 40 °C, respectively. Error bars and dashed lines show means ± SE
The estimated average cell cycle durations were 206, 202, 202, 200 and 195 min for step durations of 30, 60, 90, 120 and 150 min, respectively. With these step durations, each step durations was shorter than the length of the complete cell cycle. The estimated average cell cycle durations were 172, 157 and 162 min for step durations of 240, 480 and 720 min. For these longer step durations, multiple cell cycles happened within one step.
Effects of epigenetic mechanisms in fluctuating environment
Next we tested if epigenetic mechanisms were important for growth in fluctuating environments. Growth rates of most strains were around 2.51 mm/h with step duration of 30 min and increased to 2.70 mm/h when step duration increased to 150 min. With longer step durations, growth rate further increased to around 3.34 mm/h (Fig. 2). In a mixed model, the effect of step duration was significant (F = 383.14, numerator df = 7, denominator df = 137.7, p < 2.2 × 10−16), showing that different step durations affected the growth of the strains in fluctuating environments. Reaction norms for growth in fluctuating environments of the genotypes show that in general there was a small increase in growth rate for short-step durations until the 150-min step, and then growth rate increased faster as the strains had more time to acclimate in the 240 and 480 min steps, but growth rate plateaued after that (Fig. 2). We also tested whether the small increase in growth rate observed for the short-step durations was significant by analysing the steps from 30 to 150 min separately. In this model, the effect of duration of the step was significant (F = 18.69, numerator df = 4, denominator df = 489.01, p = 2.62 × 10−14), indicating that while the effect of duration of the step in short timescales may be small, it was likely a real effect.
Reaction norms of different classes of mutants. Lines and error bars show means ± SE, n = 5. a DNA methylation mutants. b Histone methylation mutants. c Histone deacetylation class I mutants. d Histone deacetylation class III mutants. e RNA interference mutants. f Histone demethylation and acetylation mutants
There were differences in reaction norm elevation among the different genotypes, as shown by a significant genotype effect (χ2 = 387.62, df = 1, p = 2.73 × 10−86). Differences in reaction norm elevation indicate that some mutants grew slower overall (Fig. 2), some of the mutants were undoubtedly impaired in their normal cellular metabolism and grew slower as a consequence. However, we were interested in differences in reaction norm shape, which indicated that responses to the environment were different in the different strains. There was evidence that reaction norm shapes were different as indicated by a significant step, duration × genotype interaction (χ2 = 17.18, df = 1, p = 3.4 × 10−5). Some strains responded to the environment differently, suggesting that epigenetic mechanisms affect the growth rate in fluctuating temperatures. To identify which mutants reacted differently than the control strain, we performed a pairwise ANOVA where we tested each mutant separately (Table 2).
Estimating acclimation times
Growth rate in short-term fluctuations could not be explained by the naive model of instant acclimation and was close to growth rate in constant 40 °C. Thus, we suspected that there was a lag time in acclimation response. To estimate how fast the growth rates changed, we performed temperature shift experiments, where we shifted cultures from 30 to 40 °C and vice versa and monitored how growth rate changed. In the first experiment, when cultures were shifted from 30 to 40 °C, growth rate initially dropped to around 2 mm/h and then recovered to around 2.6 mm/h (Fig. 3a). This took around 6 h, there was an increase in the growth rate around 8 h, but later the growth rate seemed to stabilise to a slightly lower value. The origin of this transient increase is unknown, it showed up earlier also in the qde-2 mutant, so it probably cannot be attributed to a random fluctuation in the growth chamber conditions. However, the overall pattern of low initial growth rate after the switch and subsequent recovery was clear. Lag time parameter, β2 ± SE, obtained from a non-linear fit was 1.38 ± 0.34 h for genotype 2489 and 0.48 ± 0:36 h for qde-2. This difference of 0.9 was suggestive but not significant (t = −1.84, df = 113, p = 0.0691). When the cultures were shifted from 40 to 30 °C, growth rate recovered to the growth rate at constant 30 °C, but this took up to 8 h (Fig. 3b). Lag time parameter, β2 ± SE, obtained via non-linear fit was 2.06 ± 0.18 h for control genotype 2489 and 3.56 ± 0.27 h for genotype qde-2. The difference of 1.49 h was significant (t = 4.57, df = 130, p = 1.11 × 10−5). These data show that temperature acclimation in Neurospora is not instantaneous but takes up to 6–8 h. Furthermore, acclimation is asymmetric; the direction of environmental change matters, as recovery from high temperature took longer than acclimation to high temperature. Acclimation times were much longer than time it took for growth chambers to equilibrate to a certain temperature (Supplementary figure S3), so temperature transfer cannot explain asymmetries in acclimation. Control and qde-2 differed in their responses, in particular the qde-2 mutant recovers slower from high temperature than the control strain (Fig. 3b). Acclimation times were also longer than estimated cell cycle times of 1.7 and 3.6 h for 30 or 40 °C, respectively. Thus, acclimation takes approximately from 2 to 5 cell cycles.
We then performed a second temperature shift experiment, where we estimated the lag times for all of the mutants. Estimates of lag parameter β2 were similar in shift from 30 to 40 °C to first experiment for the control genotype. None of the mutants had significantly different lag parameters from the control, even though some, such as hda-1 had increased variation (Fig. 4). For the set-1 mutant, the lag parameter could not be estimated as the model could not be fitted, as its growth rate did not recover from the initial depression after the transfer (Fig. S4). For the shift from 40 to 30 °C, the lags were somewhat lower than in the first lag experiment, the lag parameter β2 was 1.79 for the control. The set-1 mutant had a longer lag β2 = 3.73 ± 0.35 and difference to the control was significant (t = 3.57, df = 443, p = 4.02 × 10−4. In this experiment, the qde-2 mutant also had a longer lag time than the control (Fig. 4), but this difference was not significant (t = 1.62, df = 443, p = 0.1067). It is likely that this is because the second experiment had less statistical power. For the other mutants, lag times were not different from the control (Figs. 4 and S5). Next we will look at growth in fluctuating environment and acclimation responses of the different mutant classes in more detail.
DNA methylation
Reaction norms for growth rate in fluctuating environments of strains with deficiencies in DNA methylation are shown in Fig. 2a. The dim-2 mutant lacks a DNA methyltransferase enzyme but showed no difference to the control. Indicating that DNA methylation is not important for temperature response in Neurospora. The dmm-2 mutant is deficient in containing DNA methylation to heterochromatic regions, and as a consequence DNA methylation spreads to normally unmethylated regions in this mutant. It grew slower than the control but responded to different step durations the same way as the control. Neither mutant had any differences in acclimation (Figs. 4, S4A, S5A). This is consistent with our previous observation that neither mutant had any differences compared to control in their temperature response measured in constant temperatures (Kronholm et al. 2016).
Histone methylation
We tested three different mutants with deficiencies in histone methylation, set-1 which lacks H3K4 trimethylation, set-2 which is impaired in H3K36 methylation and set-7 which lacks H3K27 trimethylation. The growth of set-7 mutant was not different from the control (Figs. 2b, 4), but both set-1 and set-2 had lowered reaction norm elevation. Furthermore, both set-1 and set-2 differed from the control in reaction norm shape (Table 2). Both genotypes had reached their maximal growth rate already at the 240-min step, and during the short fluctuations they did not increase their growth rate as much as the control (Fig. 2b). We had previously observed that the temperature response of set-2 is altered (Kronholm et al. 2016), which may explain its response in fluctuating environments. For set-2 there were no significant differences in acclimation (Fig. 4). We evaluated if changes in reaction norm shape could be the result of this strain reaching its steady-state growth rate faster if it has to acclimate less than the control, even if acclimation happens at the same rate. We calculated the expected growth rate under the assumption of no lag from their growth rates at constant temperature, data obtained from Kronholm et al. (2016). For set-2 growth rate in 720 min step was 2.08 mm/h and this was exactly at the no lag expectation of 2.08 mm/h. Thus, for set-2 it is possible that differences in reaction norm shape reflect just lower acclimation requirement. Acclimation for set-1 took longer in the 40 to 30 °C switch (Fig. 4). However, for the 30 to 40 °C switch the model could not be fitted as it appears there is almost no acclimation happening for set-1 (Figure S4B). This could indicate that set-1 is required for acclimation to 40 °C.
Histone deacetylation
We tested two mutants of the histone deacetylase class I genes, hda-1 and hda-4, and four mutants of the histone deacetylase class III genes: nst-1, nst-2, nst-4 and nst-7. The hda mutants grew slower than the control (Fig. 2c, Table 2), but there were no significant differences in the reaction norm shape or acclimation time (Fig. 4). The hda-1 had higher acclimation time and higher variability in 30 to 40 °C acclimation, but this effect was not significant. This is probably a reflection of poor model fit, as the shape of the acclimation shows very little acclimation (Figure S4C). So it remains possible that hda-1 has some effect. For the nst mutants, nst-1 and nst-2 did not differ from the control, nst-4 had slightly lower elevation than the control but same shape and nst-7 grew very poorly, overall indicating severe problems in normal cellular functioning (Fig. 2d, Table 2). No differences in acclimation were observed for the nst mutants (Figs. 4, S4D, S5D).
RNA interference
Neurospora produces a diverse set of different small RNAs. We tested six mutants in the RNA interference pathway, including the two Dicer ribonuclease genes dcl-1, dcl-2 and their double mutant dcl-1 dcl-2, qde-1, qde-2 and qip. The growth of all other mutants except qde-2 did not differ from the control (Fig. 2e). The qde-2 mutant grew overall slower than the control, but its reaction norm shape was not different from the control (Table 2). In short-step durations, it looked like qde-2 did not increase its growth rate like the control, but this effect was not significant. In the temperature shift experiments, qde-2 acclimated slower than the control in the 40 to 30 °C shift (Figs. 3, 4, S5E), which may explain the reaction norm trend even if there was a lack of power.
Histone demethylation and acetylation
The ELP3 protein is an inferred histone acetyl transferase, while LID2 and AOF2 are inferred histone demethylases. All three mutant strains had a lower growth rate in fluctuating environments than the control (Fig. 2f), but they had the same reaction norm shape as the control (Table 2). This was expected based on our previous results, as we found no differences in the temperature response of these mutants (Kronholm et al. 2016). Furthermore, no differences in acclimation were observed for these mutants in constant temperatures (Figs. 4, S4F, S5F).
Modelling growth in fluctuating environments
The long lag times suggest a mechanistic explanation for slow growth rate in rapidly fluctuating environments. We tested this by fitting a growth model based on the observed lag times. We first fitted non-linear regressions to the lag data to estimate empirical parameters for the lag functions (Eq. (3), Table 3). Then we used those lag functions to predict the growth rates in fluctuating environments with different step durations. For the initial lag model, we calculated the predicted growth rates by integrating over the times the culture spent in the different temperatures. Comparing the predictions from the initial lag model to the entire observed data set showed that the initial lag model worked poorly (Fig. 5), particularly in short-step durations. The initial model probably failed because in short-step durations there was not enough time to acclimate completely and reach the expected growth rate. We then refined the initial model to account for partial acclimation during the short time intervals and the refined model fitted the data much better (Fig. 5). To assess how well each model fitted the data, we calculated the MSD for each predicted value from the observed data as a measure of model fit for the two different lag models, naive expectation of no lag and for growth rate in constant 40 °C. The MSD values were: 0.4 for the model with no lag, 0.31 for the initial lag model, 0.31 for the growth rate in constant 40 °C and 0.02 for the refined model. All the other models were clearly inferior compared with the refined lag model. These results indicate that the growth rate of Neurospora in fluctuating environments can be predicted to a reasonable degree if the lag functions are known.
Comparison of how well different growth models with lag fit to the observed fluctuation data of genotype 2489. For the observed data, points are means ± SE, n = 5. Red line is the growth in constant 40 °C, dashed lines and error bars show mean ± SE. Grey dashed line is predicted growth rate if there were no lag
Discussion
Growth in fluctuating environments
We investigated the stepwise fluctuations in temperature and could show that for N. crassa, fast fluctuations slowed the growth down more than expected based on growth rates under constant temperatures, and this was due to the time it took to recover from the hot temperature. If fluctuations were faster than recovery times, the fungus never reached its higher growth rate in the cooler temperature. Once the duration of fluctuation steps increased beyond the recovery time, growth in fluctuating environment sped up to or near the levels one would expect based on reaction norms obtained in constant temperatures. The periods of fluctuations used in this study were short and these kinds of fluctuations may be rare in nature. However, the important question is how does the period of environmental fluctuations scale with acclimation time? In species where acclimation time is longer, environmental fluctuations with longer periods may have similar effects as reported here. This study used large shifts in temperature that again may be rare in nature. It is therefore possible that allowing the cultures to first acclimate to intermediate temperatures would change the acclimation times. However, it is unlikely that results would change completely, for example growth rate at 35 °C is higher than growth rate at 30 °C and acclimation to stressful temperature of 40 °C is nevertheless required.
Role of epigenetic mechanisms in fluctuating environments
Our results suggest that epigenetic mechanisms have a small role in responding to fluctuating temperatures. This contrasts with our results of the role of epigenetic mechanisms in constant temperatures, where much stronger effects were observed on phenotypic plasticity (Kronholm et al. 2016). Possibly, this is because the time frame of environmental fluctuations investigated here may be faster than the normal timescale of epigenetic regulation, at least for the shortest step durations, which were well below the estimated cell cycle times. Transcriptional response to environmental changes can be rapid (Causton et al. 2001; Chechik and Koller 2009) and occur within minutes, and in yeasts, the half-life of most proteins is around 10 h, but with considerable heterogeneity among different proteins (Christiano et al. 2014). While there is evidence that some chromatin state changes occur in a circadian manner (Sahar and Sassone-Corsi 2013), and thus there was potential for epigenetic changes to occur during the fluctuations, not much is known about the time scale of epigenetic regulation in general.
We observed a reaction norm change and slower acclimation for set-1 mutant, which is deficient in histone 3 lysine 4 trimethylation, and has improper expression of certain genes (Raduwan et al. 2013). The H3K4me3 modification has been associated with 5′ regions of actively transcribed genes (Pokholok et al. 2005; Ardehali et al. 2011), and thus the absence of this histone mark may interfere with the proper activation of correct transcription response required for acclimation. The H3K4me3 mark is also required for proper expression of the circadian clock in Neurospora (Raduwan et al. 2013). The clock is affected by temperature (Crosthwaite and Heintzen 2010), and temperature changes used in this study were large enough to reset the clock (Crosthwaite and Heintzen 2010). However, in previous experiments, mutants affecting the circadian period did not show any differences in growth rate (Gardner and Feldman 1981). Thus, temperature-mediated effects of set-1 are unlikely to be mediated through the clock.
The histone mark histone 3 lysine 36 methylation, which is absent in the set-2 mutant (Adhvaryu et al. 2005), may also be important in fluctuating environments. However, no acclimation effects were detected for set-2, so some other mechanism than acclimation time has to be in play. The role of H3K36me is likely to keep transcriptionally active regions of the chromosome in open conformation (Venkatesh et al. 2012) and the mark is present in actively transcribed regions (Hampsey and Reinberg 2003; Morris et al. 2005). We have previously observed that set-2 is important in temperatures above 25 °C (Kronholm et al. 2016). If the absence of H3K36 methylation slows or otherwise interferes with the rates of transcription at high temperatures, this can explain the observation that the set-2 mutant also affects growth in fluctuating environments.
It may be surprising that we did not observe significant effect for the qde-2 mutant in fluctuating environments. In constant temperatures, we have found evidence that Neurospora ARGONAUTE homologue encoded by qde-2 is important in response to constant temperatures (Kronholm et al. 2016). We also observed this in the temperature shift experiments, where the qde-2 mutant acclimated slower than the control strain. However, it may be that this difference is not large enough to be detected when we let the strains grow in the fluctuating environments. QDE-2 protein is responsible for processing of diverse types of RNAs, such as siRNAs (Maiti et al. 2007) and microRNA-like RNAs (Lee et al. 2010) in Neurospora. Small RNAs have been linked to temperature responses in different organisms (Chen et al. 2015; Kronholm et al. 2016; Fast et al. 2017); they have the potential to act as thermoregulatory molecules as they can adopt different conformations based on temperature, and especially in prokaryotes, there are examples of RNAs acting as thermosensors (Sengupta and Garrity 2013).
Predicting growth in fluctuating environments
There has been lot of interest in predicting the performance in fluctuating environments based on reaction norms measured in constant environments (Niehaus et al. 2012; Rezende et al. 2014; Ketola and Saarinen 2015; Kingsolver et al. 2015; Kingsolver and Woods 2016; Sinclair et al. 2016 and Ketola and Krinstensen 2017). Our results show that prediction works in some circumstances; intuitively, if fluctuations are much slower than the time it takes to acclimate to a new temperature, then the performance in constant environments predicts performance in a fluctuating environment. In addition, if fluctuations happen faster than acclimation time, then performance in fluctuating environments can be predicted to some degree if acclimation times, in our case lag functions, are known. However, there are some complications that may be general. Importantly, the lag functions we estimated for N. crassa were not symmetric and the shape of the functions are likely to change when the organism has only partially acclimated to a new temperature and the temperature changes again. This is reflected in our results, our refined lag model gave the worst predictions in step durations of 60–150 min where some acclimation happened, but which was not enough time to completely acclimate. With longer step durations, predictions of the model were much better and were nearly completely in line with the observed data.
In a recent modelling study, Kingsolver and Woods (2016) introduced a general model to predict performance in fluctuating environment specifically accounting for time-dependent effects. Time-dependent effects in this model were framed in terms of heat shock protein production and degradation. It was assumed that both production and degradation happen at similar rates, but this assumption is likely to be violated in several cases, as highlighted by asymmetric acclimation responses in our data. More empirical estimates of acclimation responses are needed so that these assumptions can be relaxed in future work on performance curves.
Conclusions
Surprisingly, we did not find that epigenetic mechanisms are required for tolerating fluctuations that happen within one generation. There were some effects on acclimation time, but in these cases no memory effects seem to be involved, rather the physiological acclimation response seems to be impaired. Our results apply to within-generation effects, and possible transgenerational effects need to be investigated separately. Predicting the performance in fluctuating environments depends on the timescale of environmental fluctuations and acclimation functions of the organism in question. Increasing the complexity of the models allows taking lags into account, but empirical data are needed to parameterise the models. An added complication is that acclimation seems to be asymmetric and the shape of acclimation functions is likely to change when the temperature range of acclimation changes. These results warrant caution when using reaction norms to predict the performance in fluctuating environments. One cannot simply use data measured in constant environments and extrapolate to fluctuating environments, but by measuring acclimation functions with careful experiments, reasonably accurate models for growth in fluctuating environments can be constructed. Such experiments can also give insight into the mechanisms of acclimation.
Data archiving
Data and R scripts for analysis have been deposited in the Dryad Digital Repository, https://doi.org/10.5061/dryad.s267j33.
References
Adhvaryu KK, Morris SA, Strahl BD, Selker EU (2005) Methylation of histone H3 lysine 36 is required for normal development in Neurospora crassa. Euk/ryot Cell 4:1455–1464. http://ec.asm.org/content/4/8/1455.abstract
Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T (2011) Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J 30:2817–2828. https://doi.org/10.1038/emboj.2011.194
Arguelles JC (1997) Thermotolerance and trehalose accumulation induced by heat shock in yeast cells of Candida albicans. FEMS Microbiol Lett 146:65–71. https://doi.org/10.1111/j.1574-6968.1997.tb10172.x
Bannister AJ, Kouzarides T (2011) Regulation of chromatin by histone modifications. Cell Res 21:381–395. https://doi.org/10.1038/cr.2011.22
Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16:6–21. http://genesdev.cshlp.org/content/16/1/6.short
Bonini BM, Neves MJ, Jorge JA, Terenzi HF (1995) Effects of temperature shifts on the metabolism of trehalose in Neurospora crassa wild type and a trehalase-deficient (tre) mutant. Evidence against the participation of periplasmic trehalase in the catabolism of intracellular trehalose. Biochim Biophys Acta-Gen Subj 1245:339–347. http://www.sciencedirect.com/science/article/pii/0304416595000984
Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG et al. (2001) Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 12:323–337. http://www.molbiolcell.org/content/12/2/323.abstract
Chechik G, Koller D (2009) Timing of gene expression responses to environmental changes. J Comput Biol 16:279–290. https://doi.org/10.1089/cmb.2008.13TT
Chen J, Nolte V, Schlötterer C (2015) Temperature-related reaction norms of gene expression: regulatory architecture and functional implications. Mol Biol Evol 32:2393–2402. http://mbe.oxfordjournals.org/content/32/9/2393.abstract
Chevin LM, Lande R, Mace GM (2010) Adaptation, plasticity, and extinction in a changing environment: towards a predictive theory. PLoS Biol 8:e1000357
Christiano R, Nagaraj N, Frohlich F, Walther TC (2014) Global proteome turnover analyses of the yeasts S. cerevisiae and S. pombe. Cell Rep 9:1959–1965. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4526151/
Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L et al. (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA 103:10352–10357
Crosthwaite SK, Heintzen C (2010) Detection and response of the Neurospora crassa circadian clock to light and temperature. Fungal Biol Rev 24:114–122. http://www.sciencedirect.com/science/article/pii/S1749461310000576
Day T, Rowe L (2002) Developmental thresholds and the evolution of reaction norms for age and size at life-history transitions. Am Nat 159:338–350
de la Paz Sanchez M, Aceves-García P, Petrone E, Steckenborn S, Vega-León R, Álvarez-Buylla ER et al. (2015) The impact of Polycomb group (PcG) and Trithorax group (TrxG) epigenetic factors in plant plasticity. New Phytol 208:684–694. https://doi.org/10.1111/nph.13486
De Virgilio C, Hottiger T, Dominguez J, Boller T, Wiemken A (1994) The role of trehalose synthesis for the acquisition of thermotolerance in yeast I. Genetic evidence that trehalose is a thermoprotectant. Eur J Biochem 219:179–186. https://doi.org/10.1111/j.1432-1033.1994.tb19928.x
Deutsch CA, Tewksbury JJ, Huey RB, Sheldon KS, Ghalambor CK, Haak DC et al. (2008) Impacts of climate warming on terrestrial ectotherms across latitude. Proc Natl Acad Sci USA 105:6668–6672. http://www.pnas.org/content/105/18/6668.abstract
DeWitt TJ, Scheiner SM (2004). Phenotypic variation from single genotypes: a primer. In: DeWitt TJ, Scheiner SM (eds.), Phenotypic plasticity: functional and conceptual approaches. Oxford University Press, Inc., New York.
Fast I, Hewel C, Wester L, Schumacher J, Gebert D, Zischler H et al. (2017) Temperature-responsive miRNAs in Drosophila orchestrate adaptation to different ambient temperatures. RNA 23:1352–1364. http://rnajournal.cshlp.org/content/early/2017/06/19/rna.061119.117.abstract
Feder ME, Hofmann GE (1999) Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol 61:243–282. https://doi.org/10.1146/annurev.physiol.61.1.243. PMID: 10099689
Gardner GF, Feldman JF (1981) Temperature compensation of circadian period length in clock mutants of Neurospora crassa. Plant Physiol 68:1244–1248. http://www.plantphysiol.org/content/68/6/1244
Gaydos LJ, Wang W, Strome S (2014) H3K27me and PRC2 transmit a memory of repression across generations and during development. Science 345:1515–1518. http://www.sciencemag.org/content/345/6203/1515.abstract
Glatz A, Pilbat AM, Németh GL, Vince-Kontár K, Jósvay K, Hunya A et al. (2015) Involvement of small heat shock proteins, trehalose, and lipids in the thermal stress management in Schizosaccharomyces pombe. Cell Stress Chaperon 21:327–338. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4786532/
Hampsey M, Reinberg D (2003) Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell 113:429–432. http://www.sciencedirect.com/science/article/pii/S009286740300360X
Holm S (1979) A simple sequentially rejective multiple test procedure. Scand J Stat 6:65–70
Ingleby FC, Hunt J, Hosken DJ (2010) The role of genotype-by-environment interactions in sexual selection. J Evol Biol 23:2031–2045. https://doi.org/10.1111/j.1420-9101.2010.02080.x
Johnston IA, Dunn J (1987) Temperature acclimation and metabolism in ectotherms with particular reference to teleost fish. Symp Soc Exp Biol 41:67–93
Ketola T, Kellermann VM, Loeschcke V, Lppez-Sepulcre A, Kristensen TN (2014) Does environmental robustness play a role in fluctuating environments? Evolution 68:587–594. https://doi.org/10.1111/evo.12285
Ketola T, Kristensen TN (2017) Experimental approaches for testing if tolerance curves are useful for predicting fitness in fluctuating environments. Front Ecol Evol 5:129. https://doi.org/10.3389/fevo.2017.00129
Ketola T, Saarinen K (2015) Experimental evolution in fluctuating environments: tolerance measurements at constant temperatures incorrectly predict the ability to tolerate fluctuating temperatures. J Evol Biol 28:800–806. https://doi.org/10.1111/jeb.12606
Kingsolver JG, Diamond SE, Buckley LB (2013) Heat stress and the fitness consequences of climate change for terrestrial ectotherms. Funct Ecol 27:1415–1423. https://doi.org/10.1111/1365-2435.12145
Kingsolver JG, Higgins JK, Augustine KE (2015) Fluctuating temperatures and ectotherm growth: distinguishing non-linear and time-dependent effects. J Exp Biol 218:2218–2225. http://jeb.biologists.org/content/218/14/2218
Kingsolver JG, Ragland GJ, Diamond SE (2009) Evolution in a constant environment: thermal fluctuations and thermal sensitivity of laboratory and field populations of Manduca sexta. Evolution 63:537–541. https://doi.org/10.1111/j.1558-5646.2008.00568.x
Kingsolver JG, Woods HA (2016) Beyond thermal performance curves: Modeling time-dependent effects of thermal stress on ectotherm growth rates. Am Nat 187:283–294. https://doi.org/10.1086/684786. PMID: 26913942
Kooke R, Johannes F, Wardenaar R, Becker F, Etcheverry M, Colot V et al. (2015) Epigenetic basis of morphological variation and phenotypic plasticity in Arabidopsis thaliana. Plant Cell 27:337–348. http://www.plantcell.org/content/early/2015/02/10/tpc.114.133025.abstract
Kronholm I, Johannesson H, Ketola T (2016) Epigenetic control of phenotypic plasticity in the filamentous fungus Neurospora crassa. Genes|Genomes|Genet 6:4009–4022. http://www.g3journal.org/content/early/2016/09/29/g3.116.033860.abstract
Kuznetsova A, Brockhoff PB, Christensen RHB (2015). lmerTest: tests in linear mixed effects models. R package version 2.0-29. http://CRAN.R-project.org/package=lmerTest
Lande R (2009) Adaptation to an extraordinary environment by evolution of phenotypic plasticity and genetic assimilation. J Evol Biol 22:1435–1446
Lee HC, Li L, Gu W, Xue Z, Crosthwaite SK, Pertsemlidis A et al. (2010) Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol Cell 38:803–814. http://www.sciencedirect.com/science/article/pii/S1097276510002820
Levy O, Buckley LB, Keitt TH, Smith CD, Boateng KO, Kumar DS, et al. (2015). Resolving the life cycle alters expected impacts of climate change. Proc R Soc Lond B 282. http://rspb.royalsocietypublishing.org/content/282/1813/20150837
Luna E, Ton J (2012) The epigenetic machinery controlling transgenerational systemic acquired resistance. Plant Signal Behav 7:615–618. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3442853/
Maiti M, Lee HC, Liu Y (2007) QIP, a putative exonuclease, interacts with the Neurospora argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev 21:590–600. https://doi.org/10.1101/gad.1497607
Martegani E, Tome F, Trezzi F (1981) Timing of nuclear division cycle in Neurospora crassa. J Cell Sci 48:127–136. http://jcs.biologists.org/content/48/1/127
McCluskey K, Wiest A, Plamann M (2010) The fungal genetics stock center: a repository for 50 years of fungal genetics research. J Biosci 35:119–126
Metzenberg RL (2003) Vogel’s medium N salts: avoiding the need for ammonium nitrate. Fungal Genet Newsl 50:14
Morris SA, Shibata Y, Ki Noma, Tsukamoto Y, Warren E, Temple B et al. (2005) Histone H3 K36 methylation is associated with transcription elongation in Schizosaccharomyces pombe. Eukaryot Cell 4:1446–1454. http://ec.asm.org/content/4/8/1446.abstract
Niehaus AC, Angilletta MJ, Sears MW, Franklin CE, Wilson RS (2012) Predicting the physiological performance of ectotherms in fluctuating thermal environments. J Exp Biol 215:694–701. http://jeb.biologists.org/content/215/4/694
Nussey DH, Postma E, Gienapp P, Visser ME (2005) Selection on heritable phenotypic plasticity in a wild bird population. Science 310:304–306. http://science.sciencemag.org/content/310/5746/304
Padilla DK, Adolph SC (1996) Plastic inducible morphologies are not always adaptive: the impotance of time delays in a stochastic environment. Evol Ecol 10:105–117
Piper PW (1993) Molecular events associated with acquisition of heat tolerance by the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 11:339–355. https://doi.org/10.1111/j.1574-6976.1993.tb00005.x
Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI et al. (2005) Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122:517–527. https://doi.org/10.1016/j.cell.2005.06.026
R Core Team 2013. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org
Raduwan H, Isola AL, Belden WJ (2013) Methylation of histone H3 on lysine 4 by the lysine methyltransferase SET1 protein is needed for normal clock gene expression. J Biol Chem 288:8380–8390
Rasmann S, De Vos M, Casteel CL, Tian D, Halitschke R, Sun JY et al. (2012) Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol 158:854–863. http://www.plantphysiol.org/content/158/2/854.abstract
Rezende EL, Castañeda LE, Santos M (2014) Tolerance landscapes in thermal ecology. Funct Ecol 28:799–809. https://doi.org/10.1111/1365-2435.12268
Ryan FJ, Beadle GW, Tatum EL (1943) The tube method of measuring the growth rate of Neurospora. Am J Bot 30:784–799. http://www.jstor.org/stable/2437554
Saarinen K, Laakso J, Lindström L, Ketola T (2018) Adaptation to fluctuations in temperature by nine species of bacteria. Ecol Evol 8:2901–29. https://doi.org/10.1002/ece3.3823
Sahar S, Sassone-Corsi P (2013). The epigenetic language of circadian clocks. In: Kramer A, Merrow M (eds.), Circadian clocks. Handbook of experimental pharmacology, vol. 217. Springer, Berlin, Heidelberg.
Scheiner SM (2002) Selection experiments and the study of phenotypic plasticity. J Evol Biol 15:889–898. https://doi.org/10.1046/j.1420-9101.2002.00468.x
Schulte PM, Healy TM, Fangue NA (2011) Thermal performance curves, phenotypic plasticity, and the time scales of temperature exposure. Integr Comp Biol 51:691–702. https://doi.org/10.1093/icb/icr097
Sengupta P, Garrity P (2013) Sensing temperature. Curr Biol 23:R304–R307. http://www.sciencedirect.com/science/article/pii/S0960982213002753
Sinclair BJ, Marshall KE, Sewell MA, Levesque DL, Willett CS, Slotsbo S et al. (2016) Can we predict ectotherm responses to climate change using thermal performance curves and body temperatures. Ecol Lett 19:1372–1385. https://doi.org/10.1111/ele.12686
Turner BC, Perkins DD, Fairfield A (2001) Neurospora from natural populations: a global study. Fungal Genet Biol 32:67–92
Vasseur DA, DeLong JP, Gilbert B, Greig HS, Harley CDG, McCann KS et al. (2014) Increased temperature variation poses a greater risk to species than climate warming. Proc R Soc Lond B 281:20132612, http://rspb.royalsocietypublishing.org/content/281/1779/20132612
Venables WN, Ripley BD (2002) Modern applied statistics with S, 4th edn. Springer, New York
Venkatesh S, Smolle M, Li H, Gogol MM, Saint M, Kumar S et al. (2012) Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 489:452–455. https://doi.org/10.1038/nature11326
White B, Woodward D (1995) A simple method for making disposable race tubes. Fungal Genet Newsl 42:79
Wibowo A, Becker C, Marconi G, Durr J, Price J, Hagmann J et al. (2016) Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. eLife 5:e13546. https://doi.org/10.7554/eLife.13546
Acknowledgements
We would like to thank the Fungal Genetics Stock Center (Manhattan, Kansas), Eric Selker and Tereza Ormsby for kindly providing Neurospora strains, and Matthieu Bruneaux for critical reading of the manuscript. We would also like to thank three anonymous reviewers for their constructive comments. This research was supported by the Academy of Finland (grant no. 274769 to IK and no. 278751 to TK) and the Centre of Excellence in Biological Interactions of University of Jyväskylä.
Author contributions
IK and TK conceived the study and designed the experiments. IK performed the experiments. IK analysed the data with input from TK. IK wrote the manuscript with input from TK. Both authors gave their final approval for publication.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Kronholm, I., Ketola, T. Effects of acclimation time and epigenetic mechanisms on growth of Neurospora in fluctuating environments. Heredity 121, 327–341 (2018). https://doi.org/10.1038/s41437-018-0138-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-018-0138-2