Abstract
Studies in laboratory-based experimental evolution have demonstrated that phytoplankton species can rapidly adapt to higher temperatures. However, adaptation processes and their pace remain largely unknown under natural conditions. Here, by comparing resurrected Skeletonema marinoi strains from the Baltic Sea during the past 60 years, we show that modern S. marinoi have increased their temperature optima by 1 °C. With the increasing ability to grow in higher temperatures, growth rates in cold water decreased. Modern S. marinoi modified their valve:girdle ratio under warmer temperatures, which probably increases nutrient uptake ability. This was supported by the upregulation of several genes related to nitrate metabolism in modern strains grown under high temperatures. Our approach using resurrected strains demonstrates the adaptation potential of naturally occurring marine diatoms to increasing temperatures as global warming proceeds and exemplifies a realistic pace of evolution, which is an order of magnitude slower than estimated by experimental evolution.
Similar content being viewed by others
Main
The Anthropocene has moved the planet into a new human-mediated geological epoch1, causing a rapid loss of oceanic biodiversity on a global scale2. The increasing average temperature is one of the most evident human-induced changes. It has wide-reaching effects on many organisms2 due to the temperature dependency of biological processes3. This is especially relevant for marine organisms, as the ocean is a sink for most surplus heat4. Global sea surface temperature (SST) has risen by 0.7 °C and is projected to increase substantially by the end of the century5,6,7,8. Our study focuses on adaptation to human-induced global warming in a keystone phytoplankton species in the Baltic Sea—a region considered a ‘time machine’ for future ecosystem change due to experiencing warming levels above the global average9.
Unicellular phytoplankton, accounting for 40% of global primary production10, are essential contributors to oxygen generation, carbon sequestration and biogeochemical cycles11, and constitute the foundation of marine food webs12. Distinct thermal responses of marine phytoplankton species could lead to alterations in community composition and geographical distribution with increasing SST13. Further, productivity and diversity are expected to decline under increasing SST if species cannot shift their distribution range or adapt to the novel environment14. This may be especially true for partly isolated populations as studied here15. However, the widespread correlation between temperature optima (Topt) in phytoplankton and SST over a 150° latitudinal gradient shows that phytoplankton can adapt to different temperatures14.
The high adaptive capacity of phytoplankton has been confirmed by experimental evolution16, indicating rapid adaptation to new environments through selection on existing genetic diversity17 or de novo mutations18. Temperature adaptation to a delta of ≥4 °C has resulted in a shift either in the Topt (refs. 18,19,20,21,22,23) or the upper thermal limit18,21,22. Also, increased growth and photosynthetic rates are linked to temperature adaptation23,24. Further, analyses of differential gene expression have revealed changes in pathways related to photosynthesis and energy metabolism25,26. However, the constraints on temperature adaptation, whether due to thermodynamics6, generalist–specialist trade-offs19 or resource allocation22, remain a subject of ongoing debate. For example, temperature adaptation seems limited under low nitrogen concentrations20,27. Growth at elevated temperatures leads to an increased demand for nitrogen to sustain energetically costly repair mechanisms. This demand cannot be met under nutrient limitation because of trade-offs between allocating resources to reproduction and nutrient uptake22. Overall, experimental evolution studies have enhanced our understanding of phytoplankton evolution, but it remains uncertain whether the adaptive potential observed in laboratory experiments translates to real-world conditions.
Most experimental evolution studies do not include realistic selection pressures, natural diversity, interactions among individuals and species28, or sexual reproduction, all of which can alter the adaptation potential and/or the rate of evolution. Therefore, other approaches are required for studying adaptation under real-world global warming. This is possible through a ‘backward-in-time’ method, by resurrecting phytoplankton resting stages from seafloor sediment archives29,30,31. A previous resurrection study using phytoplankton demonstrated a shift in life cycle processes with increasing temperatures29. Analysing resurrected strains provides insights into how evolutionary processes, under a natural pace of global warming, compare to evolution occurring under simulated laboratory conditions.
With increasing SST, the general expectation is that phytoplankton cells become smaller32,33. One postulated mechanism is that size shifts allow cells to maintain the same sinking velocity with decreasing water density at high temperatures33. In addition, higher temperatures can indirectly favour smaller cells through increased resource competition and reproductive rate. Smaller cells have a larger surface:volume ratio compared with larger cells and thus a larger relative area supporting resource acquisition34, making them better competitors for resources. A shift towards smaller cells was observed in coccolithophores18 and green algae23 adapted to high temperatures. However, there is conflicting evidence for diatoms, with some species increasing and others decreasing in cell size in response to increasing temperature25. Overall, cell size decline is important to consider, as it entails the potential for far-reaching ecological consequences, including the observed productivity decline in open oceans35.
Here we study potential temperature adaptation in a natural diatom population. We compared thermal response curves, cell size and morphology, and gene expression of resurrected early-Anthropocene (1960s) strains of the key diatom species Skeletonema marinoi to strains subjected to increasing global warming (1990s and 2010s). The spring-blooming marine diatom S. marinoi is a key primary producer in the Baltic Sea36 and may periodically constitute up to 60–80% of the total biomass37. The study area in the northern Baltic Sea has been subjected to temperature increase and eutrophication during the past decades. Therefore, we expected to observe adaptation to increasing temperatures manifested as a higher Topt in modern S. marinoi (2010s). Further, we expected to observe a shift in cell size in S. marinoi from the past 60 years when grown in higher temperatures, and differential gene expression of metabolic pathways that are linked to the observed shifts in thermal reaction norms and related cell-size shifts.
The temperature optima of S. marinoi increased over the past 60 years
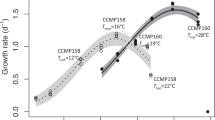
Strains of S. marinoi isolated across the past 60 years showed a shift in temperature-dependent growth and thermal performance curves (Fig. 1). The Topt of modern strains shifted by almost 1° compared with strains from the early Anthropocene (Fig. 1a,b; F2,17 = 4.98, P = 0.019). The mean Topt of 14.99 °C in the 1960s strains shifted to 15.5 °C in the 1990s strains and increased significantly to 15.88 °C in the 2010s strains (Fig. 1b). A shift of ~1.50 °C was observed in the lower temperature limit (Tmin; Fig. 1a,c; F2,17 = 15.02, P < 0.001). The strains from the 1960s showed a higher growth rate at low temperatures (Fig. 1a). Their Tmin (2.93 °C) was significantly lower compared with the 1990s and 2010s strains (3.79 °C and 4.42 °C, respectively; Fig. 1c). No differences in the upper temperature limit (Tmax) were observed between decades (Supplementary Fig. 5). The maximum growth rate (µmax) was highest in the 1990s strains at 1.01 day−1, which was 15% higher compared with the 1960s strains.
a, Mean thermal performance curves of strains from the 1960s, 1990s and 2010s (green, orange and purple, respectively). Underlying thin lines show individual performance curves of 7 strains per population. Thermal performance curves were fitted as a quadratic model. Maximum growth rate denoted by µ. b–d, Mean and confidence interval (CI) of optimum temperature (Topt) (b), lower temperature limit (Tmin) (c) and maximum growth rate (µmax) (d) were estimated for each strain by bootstrapping (n = 4 replicates per strain). Overlying strain-specific responses, the mean and CI of all strains per time point and differences between time points (lowercase grey letters) are shown.
Modern S. marinoi display a shift in cell shape in response to increased temperature. Shifts in cell size (measured as volume) along the temperature gradient from 6–22 °C were significantly different between strains of S. marinoi from different decades (Fig. 2a; F2,158 = 75.26, P < 0.001). While the size was rather similar across temperatures in strains from the 1960s and the 1990s, the modern population increased in size by 220% from low to high temperatures. Shifts in cell size affect the surface to volume ratio of the cells, which similarly showed an interaction between the temporal populations and temperature (Fig. 2b; F2,158 = 94.86, P < 0.001). The response within strains from the 1960s and 1990s was similar across temperatures, while a strong decrease (40%) occurred with increasing temperatures within the strains from the 2010s. These shifts were driven by changes in cell width (Fig. 2c,d). The cell length decreased by ~30% across strains from all decades. In contrast, shifts in cell width across temperatures were dependent on the age of strains (Fig. 2b; F2,158 = 77.00, P < 0.001). An increase of 120% was observed within the strains from the 2010s, while strains from the 1990s and 1960s showed no shift.
a–d, Volume (a), surface to volume ratio (b; S:V), width (c) and length (d) of 3 strains from the 1960s (green), 1990s (orange) and 2 strains from the 2010s (purple). Points are the mean of 50 individual cells measured per replicate. A small jitter of data points was added around the temperature values on the x axis to enhance the visibility of individual measurements. Model predictions are shown as lines.
Gene expression analysis revealed evolutionary effects on nutrient metabolism. On average, 98.2% of the RNA-seq reads per sample mapped to the S. marinoi reference genome (total size ~55 Mb). We observed no differences in mapping success between different strains (Supplementary Table 1). We compared potential differences in gene expression and found that in total, 8,280 of the 22,438 predicted genes were differentially expressed (DE) using a 5% false discovery rate (FDR) level. Approximately 76% or 6,328 of these DE genes (DEGs) received a functional annotation. When visualizing the top 500 DEGs, we observed a clear difference in expression patterns between strains from different time points (Fig. 3). The number of DEGs was higher between decades, when grown under the same temperature. The most pronounced difference was observed between the 1960s and the 2010s (4,948 ± 55 DEGs). This was significantly more than what was observed in the 1960s versus 1990s (4,021 ± 158) and the 1990s versus 2010s contrasts (3,532 ± 106) (t-test, t = 3.18, P < 0.05) (Fig. 4 and Supplementary Fig. 1).
We also observed a difference when comparing strains within time points subjected to different temperatures (Fig. 3). Between temperatures, strains from the 1960s displayed a total of 1,062 ± 138 DEGs (per strain), which is significantly less compared with that of the 1990s (1,302 ± 189) (t-test, t = 2.44, P < 0.011). Also, strains from the 2010s had significantly fewer DEGs (913 ± 240) between temperatures compared with strains from the 1990s (t-test, t = 2.44, P < 0.001) (Fig. 4). There was no difference between the 1960s and 2010s in the number of DEGs between temperatures (t-test, t = 2.44, P = 0.643). Especially in the 14 °C versus 20 °C and the 8 °C versus 14 °C contrasts, a low number of DEGs was observed, while a higher number of DEGs was observed in the 8 °C versus 20 °C contrast (Supplementary Fig. 2).
We consider genes that were uniquely DE in 2010s strains when comparing 8 °C to 20 °C to represent a portion of evolved functionality in S. marinoi (Fig. 5). There was a total of 416 such genes when including genes with log fold change (FC) values >2 or <−2. These included, for example, upregulation of thioredoxin (Acht1, Acht4), trypsin (Loc5578510) and one heat-shock gene (Hsf1) (Fig. 5). In contrast, we observed several different upregulated heat-shock genes and transcription factors (Hsf1, Hsf4, Loc4342550) in strains from the 1960s (Supplementary Table 2). When grouping genes with related functions into Gene Ontology (GO) terms, we observed a downregulation of functions within, for example, nicotinamide adenine dinucleotide phosphate (NADP) biosynthetic energy metabolism processes in strains from the 1990s and 2010s with increasing temperature (Fig. 6). This was not observed in strains from the 1960s. Also, differential gene expression in photosynthesis-related processes was observed in strains from the 1990s and 2010s but not seen in the 1960s strains. Mitotic sister chromatid cohesion (GO:0007064) was significantly upregulated in the 1990s and 2010s strains, which was not seen in the strains from the 1960s. In addition to shifts in these biological processes, we observed changes in related energy processes at the molecular function level (Supplementary Fig. 3). Changes to the photosynthesis machinery in modern strains were further supported by differential gene expression affecting cellular components within photosystem I and II (Supplementary Fig. 4).
Only biological processes that were significantly up- or downregulated in a higher temperature (8 °C→14 °C, 8 °C→20 °C, 14 °C→20 °C) are represented by a bar showing the proportion of DEGs per biological process category (gene ratio). The grey scale represents the P value from one-tailed Fisher’s exact test.
Discussion
Despite mounting evidence that thermal adaptation is possible under controlled conditions19,20,21,23, it remains uncertain how adaptation plays out under natural conditions. Multiple drivers including species interactions and abiotic environmental changes27,38 can alter the selection pressure. Adaptation may also be affected by the rate of temperature change over decades and by diurnal and annual fluctuations in temperature conditions39. This plethora of contributing factors is essential for understanding ‘real-world’ selection. Here we demonstrate that a natural phytoplankton population can increase its Topt apace with natural global warming over six decades. This provides evidence for a realistic pace of evolution of phytoplankton to global warming and several other metrics that have been under selection from global warming. We demonstrate that modern S. marinoi decreases its surface to volume ratio with increasing temperatures. This was not observed in the strains from the 1960s and 1990s. We observed shifts in how the population has altered its gene expression in relation to the increase in SST in the study area. We suggest that strains from the 1960s experienced higher temperatures as more stressful than the recent strains. This is supported by their lower growth rates in high temperatures and by a higher number of upregulated genes coding for heat-shock proteins. Heat-shock proteins are known to repair cell damage under high-temperature conditions40. The less-clear stress response in strains from 2010s exposed to above-optimum temperatures suggests that adaptation to ongoing climate change has already occurred.
In experimental evolution, temperature increase is frequently applied at the upper temperature limit of the species, posing a strong selective pressure. Here we have investigated the effect of a gradual increase in SST of ~1.5 °C in the study area since the 1960s. This entails a more subtle selection pressure compared with most experimental evolution studies that use a delta of ≥4 °C (refs. 19,20,21,23). The upper temperature tolerance limit of S. marinoi (>27 °C; Supplementary Fig. 5 and ref. 41) is never exceeded during the main growth season in this study area (Supplementary Fig. 6). This diverging selection pressure between experimental and natural evolution might partly explain why a comparable increase in Topt required more generations in our natural population compared with populations exposed to experimental evolution. For example, Chlamydomonas reinhardtii exposed for a decade to +4 °C above ambient temperature in a mesocosm study showed a 1.6 °C increase in the Topt (ref. 23). A comparable shift of 1–2 °C in Topt was observed in two diatom species grown for 200–600 generations at 4 °C above ambient temperature19. Using the climatology of the sampling area and the temperature response curves, we estimate that the observed 1 °C increase in Topt of S. marinoi required ~7,000 mitotic generations (Supplementary Fig. 6 and Supplementary Table 3). Skeletonema marinoi undergoes sexual reproduction and meiosis can be induced experimentally. However, it is unknown how often sexual events occur in nature42. Therefore, we are adhering to asexual generations in line with most laboratory evolution experiments. Also, other changing drivers in the environment (for example, light conditions and grazing), for which we have not accounted here, may have affected the population dynamics. Overall, our study demonstrates that the rate of temperature adaptation, while slower than initially estimated through experimental evolution, enables a population to adapt to ongoing global warming.
In addition, it is important to consider diurnal and seasonal temperature fluctuations, which may have contradictory effects on temperature adaptation. Diurnal fluctuations have been described to accelerate the molecular evolution of thermal tolerance in the diatom Thalassiosira pseudonana compared with constant exposure39. Further, diurnal fluctuations can lead to the evolution of plasticity43. Using our data in the framework of a reaction norm (as in ref. 44) suggests that the evolution of plasticity does not play a notable role in the temperature adaptation of modern strains (Supplementary Fig. 7). Seasonal temperature fluctuations may have a contradictory role by slowing down evolution. An increase in Topt is often accompanied by a performance trade-off, including a reduced growth rate at low temperatures19,22. We observed a higher growth capacity at low temperatures in early-Anthropocene strains and a decrease in this capacity as the optimum increased. Consequently, strains with high temperature optimum favoured in late spring can have a disadvantage during cold winter seasons. The maintenance of high phenotypic diversity in isolated strains across 60 years of selection under global warming suggests temporal fluctuation in selection pressure, which favours the maintenance of high diversity45. Moreover, strong coupling between the pelagic population and benthic resting stages could mitigate adaptation to short-term environmental fluctuations. Benthic-pelagic coupling has been suggested to result in a homogeneous population structure across the seasons in dinoflagellates in the Baltic Sea46. Strong benthic-pelagic coupling has also been observed in S. marinoi in the study area47,48. Thus, it is highly unlikely that the temperature adaptation we observed was driven by seasonal or short-term fluctuations. However, increasing winter temperatures could shift the balance in favour of strains with higher Topt. The selection for higher Topt and reduced growth capacity at low temperatures might be one of the reasons underlying the later onset of the S. marinoi spring bloom observed elsewhere49,50.
The strains isolated from the 1990s displayed the highest maximum growth rates. This may be explained by a trade-off between growth and nutrient affinity. Generally, this trade-off arises as cellular resources can be allocated to reproduction or nutrient uptake51. The nitrogen requirement of repair mechanisms such as heat-shock proteins is expected to increase during temperature adaptation. Nitrogen limitation can thus constrain adaptation to increasing temperatures20, making phytoplankton in areas with low nutrient conditions more vulnerable to global warming52. Hence, even though not directly tested here, adaptation to global warming in the population of S. marinoi in this study might have been enabled by the strong and documented eutrophication in the area53. When the limiting nutrient is present in excess, it gives individuals with higher growth a competitive advantage54. A higher growth rate is positively correlated with a higher maximum uptake velocity (Vmax), which is generally observed in the velocity-adapted diatoms. These traits are advantageous under high nutrient environments55. In our study area, eutrophication peaked in the 1990s9,56, which may explain the high growth rates in strains from this time. In agreement, a recent study from the Baltic Sea comparing populations of S. marinoi growing under different trophic states showed that eutrophication probably drives selection for faster growth57. This is further supported by the differential gene expression relating to energy metabolism and cell division (for example, GO:0046496, GO:0006739, GO:0007064). These processes are related to NADP, the central electron carrier during the light-dependent photosynthesis reactions which were most differentially expressed in strains from the 1990s and 2010s. Overall, this prompts the question of whether adaptation with ongoing global warming will be of broad applicability or if it is limited to regions subject to strong eutrophication.
Elevated temperatures tend to decrease individual cell sizes32,33 due to resource constraints and enhanced reproductive rates. However, the silica frustules of diatoms may contribute to conflicting evidence of size changes in higher temperatures25. Here we show opposing responses in size and surface:volume ratio within the same species with different Topt. While both early-Anthropocene and modern strains showed a reduced cell length under increasing temperatures, modern strains with higher Topt showed an increase in cell width. This is in line with shifts in cell size associated with temperature evolution in the diatom Thalassiosira pseudonana58. Diatoms respond to resource limitation by adjusting the valve:girdle ratio. This ratio is relevant for nutrient acquisition, as the silica frustules restrict nutrient uptake at the elongated girdle band (length) while facilitating it at the circular valves (width) that are equipped with punctae. With increasing valve:girdle ratio, the ratio of the surface area of the valve to the volume of the cell also increased58 (Supplementary Fig. 8) and the cell shape changed (Supplementary Fig. 9). Overall, the counterintuitive increase in size with increasing temperature adaptation is probably driven by an increased need for resources. We also found support for this higher demand for nutrients in the gene expression of modern S. marinoi. Among the upregulated genes in high temperature, we observed several with functions related to trypsin metabolism, which is a known regulator of N:P stoichiometric homoeostasis in phytoplankton59. The expression of trypsin in diatoms is especially responsive to shifts in the environment60. Here, it may be the key to fulfil the higher demand for nitrate when temperatures have increased and the competition for this resource simultaneously intensifies. Further, two thioredoxins, which are central regulators of CO2 fixation and nitrogen in chloroplasts61, were highly upregulated in strains from the 2010s. This suggests some modification of the nutrient acquisition and photosynthesis machinery in modern S. marinoi strains.
Conclusions
We conclude that S. marinoi in the Baltic Sea has adapted to ongoing global warming in the past 60 years. Our gene expression data and the observed shifts in cell morphology support earlier experimental evolution studies showing that nutrient conditions have the potential to affect adaptation to temperature increases in phytoplankton. In agreement, we also observed a trade-off between high growth at warm versus cold temperatures. However, the number of generations required by the natural population to reach the same evolutionary change was an order of magnitude greater. Overall, the underlying mechanisms of evolutionary change can be well understood in experimental studies, but the estimation of the rate of evolution requires the study of natural communities.
Methods
Model organism
The spring bloom in the northeast Baltic Sea is often dominated by the marine diatom S. marinoi which serves as an important food source for zooplankton37. This model species has been extensively studied in terms of biogeography, physiology and genetic variation across space and time in temperate areas of the world62,63. It is known to form resting stages that sink to the seafloor when the bloom phase is over towards the end of the spring bloom64. This ‘biological archive’ may contain up to ~57,000 S. marinoi cells per gram sediment64, with the potential to reveal the diversity and ecophysiology of past populations.
Study area, sediment sampling and age modelling
The Småholmen station (60.24° N, 22.04° E) close to Haverö in the Archipelago Sea is 20 m deep and has been seasonally anoxic or hypoxic at the seafloor for at least seven decades65. The low oxygen conditions and lack of bioturbation have resulted in the formation of laminated sediment at this site. Such conditions are optimal for preserving the chronology of sedimented cells. Historical data show that the average spring temperature (air) in the Archipelago Sea was stable at ~2 °C from the 1880s until the 1970s. Since the mid-1970s, it has increased by ~0.5 °C per decade66. An increase in the SST has been observed close to our study site (Seili monitoring station located <3 km from Småholmen). In April, when the spring bloom reaches its peak, SST has increased by ~2.5 °C since the early 1980s (Supplementary Fig. 10).
Sediment cores were retrieved in April 2020 using an HTH Kajak surface sediment gravity corer67 at the Småholmen station. After core retrieval, the core tube containing undisturbed sediment profiles was attached to a stand on-site and carefully sliced into 2 cm subsamples. The outer edge (3 cm) of the entire sediment core was discarded to avoid cross-contamination between different depth layers due to smearing along the outer edge of the core. The subsamples were stored in the dark at 8 °C until further processing. An age–depth model for the sediment core was constructed with the Undatable software68 using an xfactor of 0.1 and 15% bootstrapping (Supplementary Fig. 11). The xfactor value determines the sediment accumulation rate uncertainty between consecutive age–depth constraints and the bootstrapping function randomly removes a selected percentage of the age–depth constraints in each run of the age–depth simulation68. Age constraints for the modelling procedure were obtained through loss on ignition (LOI) correlation against previous studies in the same location47. LOI was determined at 2 cm intervals by drying the subsamples at 105 °C for 16 h and ashing at 550 °C for 2 h. The LOI correlation was further verified by 137Cs dating of a replicate sediment core retrieved for this study (Supplementary Fig. 12). For this, 137Cs activity of the untreated 2-cm-thick sediment slices was determined by gamma spectrometry using a BrightSpec bMCA-USB pulse height analyser coupled to a well-type NaI(Tl) detector69. In the northern hemisphere, 137Cs contamination in sediments is mainly derived from the Chernobyl nuclear power plant accident in 1986 and atmospheric nuclear weapons testing in the early 1960s70. Therefore, the 137Cs dating of Baltic Sea sediments is based on the recognition of these two horizons in the sediment profiles.
Resurrection of clonal cultures
Resurrection was initiated by mixing 0.5 g of sediment from different layers with each 50 ml of filtered (0.22 μm) seawater with f/2+Si medium71 in June 2020. The sediment was from the 2–4 cm, 28–30 cm and 46–48 cm layers corresponding to median age ± 1σ error estimates of 2017–2019–2020 (referred to as ~2010s/modern), 1985–1992–2001 (~1990s) and 1957–1963–1969 (~1960s/early Anthropocene), respectively (Supplementary Fig. 8). The sediment ‘slurries’ were distributed on 24-well NUNC plates and incubated at 8 °C and 40 μmol photons m−2 s−1 (12 h:12 h light:dark cycle). When vegetative growth emerged (after 2–4 weeks), single chains of S. marinoi were isolated using a micropipette under an inverted light microscope (Nikon Diaphot 300). One chain per well was isolated to a new well to minimize the probability of isolating the same clone twice. As previous population genetic studies on S. marinoi have shown, the genotypic diversity of S. marinoi is extremely high72. Thus, it is highly unlikely to ever find the same genotype (using, for example, microsatellite markers) of this species in natural conditions when sampled over time or across space. Therefore, we considered that the strains included in this study were all different genotypes and consisted of a small percentage of the population’s total intraspecific diversity. The isolated strains can theoretically stem from different seasons, as each layer contains resting stages across seasons from 2–5 years. We confirmed that the strains belonged to the species S. marinoi by sequencing the V4 region of the 18S ribosomal RNA gene (for more details see Supplementary Information, ‘Genetic identification of S. marinoi’). After 1–2 weeks, the entire volume in each well was transferred to 40 ml culture flasks containing 10 ml of f/2+Si media. After further growth for 7 days, the total volume was increased to 40 ml, and clonal cultures were maintained in the same conditions as described above. The resurrection rate varied between 44 and 58% (isolated cells that made it to a stable culture). The rates for the 1960s, 1990s and 2010s were 52%, 58% and 44%, respectively.
Experiments
To test whether S. marinoi has adapted to an increase in temperature during the past ~60 years, we assayed seven strains per time point (1960s, 1990s, 2010s) in +6 °C, +8 °C, +10 °C, +14 °C, +18 °C, +20 °C, +22 °C and +26 °C in natural seawater (6 PSU, 0.22 μm filtered, with f/4+Si) and 100 μmol photons m−2 s−1 (12 h:12 h light:dark cycle). Due to technical limitations, each temperature was assessed in a separate experiment in a randomized order (Supplementary Table 4). Randomization minimizes the influence of measurement timing on different temperature treatments by ensuring that the timing effect is evenly distributed across the temperature range. Before each experimental start, strains were acclimated to f/4+Si media for 1 week. The experiment was started by transferring cells to new media, reaching a start concentration of 10,000 cells per ml. To avoid dilution of nutrients, the maximum inoculum was 5 ml (10% of the total volume). The growth of each strain (four replicates) was monitored daily by measuring the in vivo fluorescence of chlorophyll a in a 300 μl subsample on a 96-well plate (PerkinElmer, IsoPlate 96F) using a spectrophotometer (Tecan, Infinite 200 Pro with the software Magellan for Tecan Infinite Pro v.1) until the stationary phase was reached. The excitation wavelength was set to 425 nm and the emission wavelength to 680 nm. To account for uneven distribution of cells, nine positions were measured in each well. The cells were not dark-adapted, but to mitigate potential confounding effects of light adaptation during the diurnal light–dark cycle, measurements were consistently taken at the same time. At the end of the experiment, 1 ml subsamples were fixed with acidic Lugol’s solution for later estimation of cell size. Three randomly chosen strains from each time point were consistently sampled for RNA when the stationary phase was reached (Supplementary Table 5). About 25–35 ml of cell culture was centrifuged for 30 min at 4 °C at 3,900 × g (Eppendorf, 5810R). The cell pellet was resuspended in 400 μl of TRIzol (Invitrogen) and incubated for 2 min at 60 °C until completely dissolved. The cells were immediately stored at −80 °C until RNA extraction.
Thermal performance curves based on growth
Growth was calculated by fitting a linear model (‘easy_linear’) to blank-corrected fluorescence values using the ‘growthrate’ package73 in R (v.4.3.0)74 and R Studio (v.2023.9.0.463)75. We excluded all replicates that showed negative growth (all replicates in 26 °C, 1960_05/06 in 22 °C) or unstable growth (maximum fluorescence <2,000) before fitting the growth model. For the samples with negative growth, the growth rate was set to zero for further analysis. Strain 2010_13 was potentially contaminated and was excluded from all downstream analyses. The calculated growth rates (total n of 216, 222 and 184 for the 1960s,1990s and 2010s, respectively) were plotted against temperature and thermal performance curves fitted using the ‘rTPC’ package24. The best-fitting model was chosen using the mean growth rate per temperature for each time point (1960s, 1990s, 2010s). Based on the Akaike information criterion (AIC) and the Bayesian information criterion (BIC), the best-fitting model was quadratic (models included: beta_2012, Gaussian, quadratic, Thomas_2012, Thomas_2017, Weinbull_1995). For each strain, confidence intervals of estimated parameters were produced by bootstrapping (maximum of 999 iterations). Bootstrapping simulated datasets from the existing data (including all four replicates) by sampling with replacement. Differences in estimated parameters such as Topt, Tmin and µmax between strains from different time points were tested using a Tukey’s HSD (honestly significant difference, two-sided). The Tmax was excluded as a primary response since (1) S. marinoi rarely occurs in the region during summer when the temperature conditions may reach levels close to or above their Tmax and (2) our attempt to capture the decline in growth rates at elevated temperatures was partly unsuccessful, leading to high uncertainty in the Tmax estimates.
Estimating cell size
We measured cell size for a subset of three strains from each time point. Lugol-fixed samples were settled for 15 min in a Sedgewick–Rafter counting chamber. Using an inverted Olympus IX 51 microscope and an Olympus U-CMAD3 digital camera, a minimum of 10 pictures per sample were taken under ×200 magnification. The width and height of five cells were measured from each picture (Olympus cellSens Dimension imaging software v.3.2). The cells were chosen randomly and only one cell per chain was measured to include the natural variability present between the chains. The silicaceous marginal processes connecting the cells were not included in the width measurements as it was not always possible to determine where the process of the next cell started. For each cell, we calculated the size as biovolume and the surface area following the formula given in ref. 76. From this, we calculated the ratio of surface area to volume (S:V). We analysed differences in volume, S:V ratio, width and length using a linear mixed effect model (LME) including genotypes nested by age of the population as a random effect lme (Volume/SV/Width/Length ~Age+temp+Age:temp+(1|Genotype:Age)). The models were fitted using the ‘lme4’ package77 in R and visualized using ‘lmerTest’78 and ‘ggplot2’79.
RNA extraction and sequencing
RNA was extracted with the RNeasy plant mini kit (Qiagen) according to manufacturer instructions with some modifications. TRIzol was removed from samples thawed on ice before extraction by centrifuging for 10 min at 8 °C at 4,500 × g. The cell pellet was washed once with nuclease-free water before proceeding with the extraction. RLT lysis buffer was added to the washed cell pellet without any additional mechanical lysis steps before proceeding with the extraction protocol according to manufacturer instructions. The quality of the RNA extracts was ensured using an Agilent Bioanalyzer 2100. Sequencing was performed on samples from the 8 °C, 14 °C and 20 °C experiments with three of the biological replicates per strain, except in cases of insufficient RNA yield (Supplementary Table 5). The library was prepared according to the Illumina Stranded mRNA library preparation protocol using 100 ng of RNA per sample as the starting material. Stranded mRNA sequencing was performed using Illumina NovaSeq 6000 (S4 v.1.5), producing ~90 million paired-end reads (2 × 150 bp) per sample.
RNA-seq data analysis
The quality of read pairs from all 60 samples was checked using FastQC (v.0.11.8)80. Only light read trimming from the ‘3 ends was conducted using Trimmomatic (TRAILING:20) to ensure a minimum (≥20) Phred33 score also towards the end of each sequence where the base quality normally drops in Illumina sequences. Since read trimming may lead to short reads and subsequent spurious alignment to the reference genome downstream, we required a minimum sequence length of 50 after trimming (MINLEN:50)81. Reads were aligned to the S. marinoi reference genome (https://zenodo.org/records/7786015) using BWA (v.0.7.17)82 (Supplementary Table 1). The resulting ‘.bam’ files were sorted and indexed using Samtools (v.1.16.1)83. Count tables for individual samples were created using Samtools, resulting in several mapped reads per predicted gene. The model fitting was conducted in edgeR84. First, the count tables were filtered using the count per million (CPM) so that only genes with at least one CPM in at least three samples were retained. The count data were normalized using a set of normalization factors (one for each sample) to eliminate composition biases between libraries. We explored the expression profiles of individual samples more closely by generating mean-difference plots (Supplementary Fig. 13). Before the differential gene expression analysis was conducted, we estimated the dispersion (Supplementary Fig. 14) and accounted for gene-specific variability from both biological and technical sources by fitting the actual model with the glmQLFit function (Supplementary Fig. 15). We calculated the leading FC by taking the root mean square of the largest 500 log2FC between pairwise samples to explore general patterns in the entire data set. For the analysis of differential gene expression, we defined different contrasts of interest. First, we analysed differences in gene expression of each strain across three temperatures (8 °C, 14 °C and 20 °C). Second, we assessed whether the mean change in gene expression across temperatures was comparable between the 1960s, 1990s and 2010s. To test whether any of the contrasts were significant (controlling for the gene-level FDR), we performed a stage-wise testing procedure in stageR85. This consisted of a screening stage and a confirmation stage conducted as in ref. 86. In total, 102 genes that were identified as significant in the screening stage were removed after the confirmation stage. We continued the analyses with 8,280 genes that displayed significant P values after FDR adjustment (alpha level 5%; Supplementary Fig. 1). Differences in the number of DEGs between decades and across temperatures were analysed using a Student’s t-test (two-sided).
The functional annotation of the S. marinoi genome was conducted using different tools. Briefly, we used BLAST+ (v.2.13.0)87 to run sequence similarity blastp searches of all S. marinoi proteins annotated in the genome against the Swissprot database. We retained the best hit using a maximum e-value limit of 1 × 10−6. InterProScan (5.55–88.0)88 was used to acquire GO terms and Pfam domains. In addition, we obtained KEGG pathway annotations using the web version of KofamKOALA89. For a gene to be considered functionally annotated, we required hits in the SwissProt database and that it received at least one assignment to Pfam domains, GO and/or KEGG annotation. GO enrichment was done with TopGO (over-representation analysis)90 and revealed the biological processes, molecular functions and cellular components the DEGs were linked to. We conducted GO enrichment tests for genes that were up- or downregulated in high temperatures based on the mean response. We used REVIGO91 to remove redundant GO terms.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All RNA sequence data produced in this study can be accessed through the National Center for Biotechnology Center (NCBI) under the BioProject code PRJNA1016074 and Sequence Read Archives (SRA) SRR26046577–SRR26046596. All S. marinoi strains are publicly available from the authors. All other experimental data are available via Zenodo at https://doi.org/10.5281/zenodo.10731675 (ref. 92). The Swissprot database (https://www.ebi.ac.uk/uniprot/download-center) was downloaded on 3 April 2023. We used the KofamKOALA version of 3 April 2023 (KEGG release 106.0).
References
Waters, C. N. et al. The Anthropocene is functionally and stratigraphically distinct from the Holocene. Science 351, aad2622 (2016).
McCauley, D. J. et al. Marine defaunation: animal loss in the global ocean. Science 347, 1255641 (2015).
Mundim, K. C., Baraldi, S., Machado, H. G. & Vieira, F. M. C. Temperature coefficient (Q10) and its applications in biological systems: beyond the Arrhenius theory. Ecol. Modell. 431, 109127 (2020).
Hoegh-Guldberg, O. & Bruno, J. F. The impact of climate change on the worlds marine ecosystems. Science 328, 1523–1528 (2010).
Boyce, D. G., Lewis, M. R. & Worm, B. Global phytoplankton decline over the past century. Nature 466, 591–596 (2010).
Pörtner, H.-O. et al. in Climate Change 2014: Impacts, Adaptation, and Vulnerability (eds Field, C. B. et al.) Ch. 6 (Cambridge Univ. Press, 2014).
Huang, J. et al. Recently amplified Arctic warming has contributed to a continual global warming trend. Nat. Clim. Change 7, 875–879 (2017).
Markus Meier, H. E. et al. Climate change in the Baltic Sea region: a summary. Earth Syst. Dyn. 13, 457–593 (2022).
Reusch, T. B. H. et al. The Baltic Sea as a time machine for the future coastal ocean. Sci. Adv. 4, eaar8195 (2018).
Falkowski, P. G. The role of phytoplankton photosynthesis in global biogeochemical cycles. Photosynth. Res. 39, 235–258 (1994).
Basu, S. & Mackey, K. R. M. Phytoplankton as key mediators of the biological carbon pump: their responses to a changing climate. Sustainability 10, 869 (2018).
Cloern, J. E. Phytoplankton bloom dynamics in coastal ecosystems: a review with some general lessons from sustained investigations of San Francisco Bay, California. Rev. Geophys. 34, 127–168 (1996).
Anderson, S. I., Barton, A. D., Clayton, S., Dutkiewicz, S. & Rynearson, T. A. Marine phytoplankton functional types exhibit diverse responses to thermal change. Nat. Commun. 12, 6413 (2021).
Thomas, M. K., Kremer, C. T., Klausmeier, C. A. & Litchman, E. A global pattern of thermal adaptation in marine phytoplankton. Science 338, 1085–1088 (2012).
Sjöqvist, C., Godhe, A., Jonsson, P. R., Sundqvist, L. & Kremp, A. Local adaptation and oceanographic connectivity patterns explain genetic differentiation of a marine diatom across the North Sea–Baltic Sea salinity gradient. Mol. Ecol. 24, 2871–2885 (2015).
Reusch, T. B. H. & Boyd, P. W. Experimental evolution meets marine phytoplankton. Evolution 67, 1849–1859 (2013).
Lohbeck, K. T., Riebesell, U. & Reusch, T. B. H. Adaptive evolution of a key phytoplankton species to ocean acidification. Nat. Geosci. 5, 346–351 (2012).
Schluter, L. et al. Adaptation of a globally important coccolithophore to ocean warming and acidification. Nat. Clim. Change 4, 1024–1030 (2014).
Jin, P. & Agustí, S. Fast adaptation of tropical diatoms to increased warming with trade-offs. Sci. Rep. 8, 17771 (2018).
Aranguren-Gassis, M., Kremer, C. T., Klausmeier, C. A. & Litchman, E. Nitrogen limitation inhibits marine diatom adaptation to high temperatures. Ecol. Lett. 22, 1860–1869 (2019).
Listmann, L., LeRoch, M., Schlueter, L., Thomas, M. K. & Reusch, T. B. H. Swift thermal reaction norm evolution in a key marine phytoplankton species. Evol. Appl. 9, 1156–1164 (2016).
O’Donnell, D. R. et al. Rapid thermal adaptation in a marine diatom reveals constraints and trade-offs. Glob. Change Biol. 24, 4554–4565 (2018).
Schaum, C. E. et al. Adaptation of phytoplankton to a decade of experimental warming linked to increased photosynthesis. Nat. Ecol. Evol. 1, 94 (2017).
Padfield, D., Yvon-Durocher, Ge., Buckling, A., Jennings, S. & Yvon-Durocher, Ga. Rapid evolution of metabolic traits explains thermal adaptation in phytoplankton. Ecol. Lett. 19, 133–142 (2016).
Liang, Y., Koester, J. A., Liefer, J. D., Irwin, A. J. & Finkel, Z. V. Molecular mechanisms of temperature acclimation and adaptation in marine diatoms. ISME J. 13, 2415–2425 (2019).
Kontopoulos, D. G. et al. Phytoplankton thermal responses adapt in the absence of hard thermodynamic constraints. Evolution 74, 775–790 (2020).
Aranguren-Gassis, M. & Litchman, E. Thermal performance of marine diatoms under contrasting nitrate availability. J. Plankton Res. 42, 680–688 (2020).
Scheinin, M., Riebesell, U., Rynearson, T. A., Lohbeck, K. T. & Collins, S. Experimental evolution gone wild. J. R. Soc. Interface 12, 20150056 (2015).
Hinners, J., Kremp, A. & Hense, I. Evolution in temperature-dependent phytoplankton traits revealed from a sediment archive: do reaction norms tell the whole story? Proc. R. Soc. B 284, 20171888 (2017).
Härnström, K., Ellegaard, M., Andersen, T. J. & Godhe, A. Hundred years of genetic structure in a sediment revived diatom population. Proc. Natl Acad. Sci. USA 108, 4252–4257 (2011).
Yousey, A. M. et al. Resurrected ‘ancient’ Daphnia genotypes show reduced thermal stress tolerance compared to modern descendants. R. Soc. Open Sci. 5, 5172193 (2018).
Sommer, U., Peter, K. H., Genitsaris, S. & Moustaka-Gouni, M. Do marine phytoplankton follow Bergmann’s rule sensu lato? Biol. Rev. 92, 1011–1026 (2017).
Zohary, T., Fishbein, T., Shlichter, M. & Naselli-Flores, L. Larger cell or colony size in winter, smaller in summer – a pattern shared by many species of Lake Kinneret phytoplankton. Inland Waters 7, 200–209 (2017).
Aksnes, D. L. & Egge, J. K. A theoretical model for nutrient uptake. Mar. Ecol. Prog. Ser. 70, 65–72 (1991).
Boyce, D. G. & Worm, B. Patterns and ecological implications of historical marine phytoplankton change. Mar. Ecol. Prog. Ser. 534, 251–272 (2015).
Wasmund, N. et al. Extension of the growing season of phytoplankton in the western Baltic Sea in response to climate change. Mar. Ecol. Prog. Ser. 622, 1–16 (2019).
Jochem, F. Distribution and importance of autotrophic ultraplankton in a boreal inshore area (Kiel Bight, Western Baltic). Mar. Ecol. Prog. Ser. 53, 153–168 (1989).
Jin, P. et al. Increased genetic diversity loss and genetic differentiation in a model marine diatom adapted to ocean warming compared to high CO2. ISME J. 16, 2587–2598 (2022).
Schaum, C. E., Buckling, A., Smirnoff, N., Studholme, D. J. & Yvon-Durocher, G. Environmental fluctuations accelerate molecular evolution of thermal tolerance in a marine diatom. Nat. Commun. 9, 1719 (2018).
Macario, A. J. L. & de Macario, E. C. Molecular chaperones: multiple functions, pathologies, and potential applications. Front. Biosci. 12, 2588–2600 (2007).
Anderson, S. I. & Rynearson, T. A. Variability approaching the thermal limits can drive diatom community dynamics. Limnol. Oceanogr. 65, 1961–1973 (2020).
Godhe, A., Kremp, A. & Montresor, M. Genetic and microscopic evidence for sexual reproduction in the centric diatom Skeletonema marinoi. Protist 165, 401–416 (2014).
Schaum, C. E., Rost, B. & Collins, S. Environmental stability affects phenotypic evolution in a globally distributed marine picoplankton. ISME J. 10, 75–84 (2016).
Stoks, R., Govaert, L., Pauwels, K., Jansen, B. & De Meester, L. Resurrecting complexity: the interplay of plasticity and rapid evolution in the multiple trait response to strong changes in predation pressure in the water flea Daphnia magna. Ecol. Lett. 19, 180–190 (2016).
Gsell, A. S. et al. Genotype-by-temperature interactions may help to maintain clonal diversity in Asterionella formosa (Bacillariophyceae). J. Phycol. 48, 1197–1208 (2012).
Jerney, J. et al. Seasonal genotype dynamics of a marine dinoflagellate: pelagic populations are homogeneous and as diverse as benthic seed banks. Mol. Ecol. 31, 512–528 (2022).
Godhe, A. & Härnström, K. Linking the planktonic and benthic habitat: genetic structure of the marine diatom Skeletonema marinoi. Mol. Ecol. 19, 4478–4490 (2010).
Sefbom, J. et al. A planktonic diatom displays genetic structure over small spatial scales. Environ. Microbiol. 20, 2783–2795 (2018).
Borkman, D. G. & Smayda, T. Multidecadal (1959–1997) changes in Skeletonema abundance and seasonal bloom patterns in Narragansett Bay, Rhode Island, USA. J. Sea Res. 61, 84–94 (2009).
Lundsor, E. et al. Changes in phytoplankton community structure over a century in relation to environmental factors. J. Plankton Res. 44, 866–883 (2022).
Klausmeier, C. A., Litchman, E., Daufreshna, T. & Levin, S. A. Optimal nitrogen-to-phosphorus stoichiometry of phytoplankton. Nature 429, 171–174 (2004).
Thomas, M. K. et al. Temperature–nutrient interactions exacerbate sensitivity to warming in phytoplankton. Glob. Change Biol. 23, 3269–3280 (2017).
Voss, M. et al. History and scenarios of future development of Baltic Sea eutrophication. Estuar. Coast. Shelf Sci. 92, 307–322 (2011).
Litchman, E., Klausmeier, C. A., Schofield, O. M. & Falkowski, P. G. The role of functional traits and trade-offs in structuring phytoplankton communities: scaling from cellular to ecosystem level. Ecol. Lett. 10, 1170–1181 (2007).
Sommer, U. The paradox of the plankton: fluctuations of phosphorus availability maintain diversity of phytoplankton in flow-through cultures. Limnol. Oceanogr. 29, 633–636 (1984).
Gustafsson, B. G. et al. Reconstructing the development of Baltic Sea eutrophication 1850–2006. Ambio 41, 534–548 (2012).
Olofsson, M., Almen, A. K., Jaatinen, K. & Scheinin, M. Temporal escape-adaptation to eutrophication by Skeletonema marinoi. FEMS Microbiol. Lett. 369, fnac011 (2022).
O’Donnell, D. R., Beery, S. M. & Litchman, E. Temperature-dependent evolution of cell morphology and carbon and nutrient content in a marine diatom. Limnol. Oceanogr. 66, 4334–4346 (2021).
You, Y. et al. Trypsin is a coordinate regulator of N and P nutrients in marine phytoplankton. Nat. Commun. 13, 4022 (2022).
You, Y., Sun, X. & Lin, S. An ancient enzyme finds a new home: prevalence and neofunctionalization of trypsin in marine phytoplankton. J. Phycol. 59, 152–166 (2023).
Kikutani, S. et al. Redox regulation of carbonic anhydrases via thioredoxin in chloroplast of the marine diatom Phaeodactylum. J. Biol. Chem. 287, 20689–20700 (2012).
Kooistra, W. H. C. F. et al. Global diversity and biogeography of Skeletonema species (Bacillariophyta). Protist 159, 177–193 (2008).
Saravanan, V. & Godhe, A. Genetic heterogeneity and physiological variation among seasonally separated clones of Skeletonema marinoi (Bacillariophyceae) in the Gullmar Fjord, Sweden. Eur. J. Phycol. 45, 177–190 (2010).
McQuoid, M. R., Godhe, A. & Nordberg, K. Viability of phytoplankton resting stages in the sediments of a coastal Swedish fjord. Eur. J. Phycol. 37, 191–201 (2002).
Jokinen, S. A. et al. A 1500-year multiproxy record of coastal hypoxia from the northern Baltic Sea indicates unprecedented deoxygenation over the 20th century. Biogeosciences 15, 3975–4001 (2018).
Laakso, L. et al. 100 years of atmospheric and marine observations at the Finnish Utö Island in the Baltic Sea. Ocean Sci. 14, 617–632 (2018).
Renberg, I. & Hansson, H. The HTH sediment corer. J. Paleolimnol. 40, 655–659 (2008).
Lougheed, B. C. & Obrochta, S. P. A rapid, deterministic age-depth modeling routine for geological sequences with inherent depth uncertainty. Paleoceanogr. Paleoclimatol. 34, 122–133 (2019).
Ojala, A. E. K., Luoto, T. P. & Virtasalo, J. J. Establishing a high-resolution surface sediment chronology with multiple dating methods – testing 137Cs determination with Nurmijärvi clastic-biogenic varves. Quat. Geochronol. 37, 32–41 (2017).
Kotilainen, A. T., Kotilainen, M. M., Vartti, V. P., Hutri, K. L. & Virtasalo, J. J. Chernobyl still with us: 137Caesium activity contents in seabed sediments from the Gulf of Bothnia, northern Baltic Sea. Mar. Pollut. Bull. 172, 112924 (2021).
Guillard, R. R. L. in Culture of Marine Invertebrate Animals (eds Smith, W. L. & Chanley, M. H.) 29–60 (Springer, 1975).
Sassenhagen, I., Erdner, D. L., Lougheed, B. C., Richlen, M. L. & Sjöqvist, C. Estimating genotypic richness and proportion of identical multi-locus genotypes in aquatic microalgal populations. J. Plankton Res. 44, 559–572 (2022).
Petzoldt, T. growthrates: Estimate Growth Rates from Experimental Data. R package version 0.8.4. GitHub https://github.com/tpetzoldt/growthrates (2022).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2022).
RStudio Team. RStudio: Integrated Development Environment for R. RStudio, PBC http://www.rstudio.com/ (2020).
Hillebrand, H., Dürselen, C.-D., Kirschtel, D., Pollingher, U. & Zohary, T. Biovolume calculation for pelagic and benthic microalgae. J. Phycol. 424, 403–424 (1999).
Douglas, B., Mächler, M., Bolker, B. & Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2015).
Kuznetsova, A., Brockhoff, P. B. & Christensen, R. H. B. lmerTest package: tests in linear mixed effects models. J. Stat. Softw. 82, 1–26 (2017).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2009).
Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Babraham Bioinformatics https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010).
Williams, J. L., Kendall, B. E. & Levine, J. M. Experimental landscapes. Science 353, 482–485 (2016).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2009).
Van den Berge, K., Soneson, C., Robinson, M. D. & Clement, L. stageR: a general stage-wise method for controlling the gene-level false discovery rate in differential expression and differential transcript usage. Genome Biol. 18, 151 (2017).
Pinseel, E. et al. Strain-specific transcriptional responses overshadow salinity effects in a marine diatom sampled along the Baltic Sea salinity cline. ISME J. 16, 1776–1787 (2022).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Jones, P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014).
Aramaki, T. et al. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36, 2251–2252 (2020).
Alexa, A. & Rahnenfuhrer, J. topGO: Enrichment Analysis for Gene Ontology. R package version 2.52.0. Bioconductor https://bioconductor.org/packages/topGO (2023).
Supek, F., Bošnjak, M., Škunca, N. & Šmuc, T. Revigo summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6, e21800 (2011).
Hattich, G. S. I. et al. Temperature optima of a natural diatom population increases as global warming proceeds. Zenodo https://doi.org/10.5281/zenodo.10731675 (2024).
Acknowledgements
This research was funded by the Academy of Finland (grant number 321609) (C.S.), the Swedish Cultural Foundation (grant number 176762) (C.S.), the European Regional Development Fund and the programme Mobilitas Pluss (MOBTP160) (M.S., M.M.), the Estonian Research Council (grant PSG735) (S.S.), the Finnish Society of Sciences and Letters (G.S.I.H.), the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation; HA 9696/1-1) (G.S.I.H.) and the Åbo Akademi University Foundation (C.S.). This study utilized research infrastructure as part of the FINMARI consortium (Finnish Marine Research Infrastructure network) and was supported by the molecular lab at Husö Biological Station, Finnish Functional Genomics Centre, Turku Bioscience, Åbo Akademi University and the University of Turku. Open access fees were covered by Gösta Branders research fund, Åbo Akademi Research Foundation. We also acknowledge CSC – IT Center for Science, Finland, for computational resources; K. Künnis-Beres, K. Pärt and L. Lattu for the measurement of cell sizes; P. Kallio for support with the spectrophotometer; and K. Ramesh for proofreading the paper.
Author information
Authors and Affiliations
Contributions
C.S. conceived the presented idea. Sediment cores were taken by C.S., M.G. and S.J. Sediment cores were dated by S.J. and M.G. M.G., C.S., G.S.I.H. and S.S. designed the experiment. Experimental work was carried out by M.G., J.H., N.J., M.S. and M.M. Cell-size measurements were conducted by S.S. C.S., G.S.I.H. and S.J. analysed the data. G.S.I.H. and C.S. wrote the paper, and all other authors revised the paper and gave final approval for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Climate Change thanks Peng Jin, Daniel R. O’Donnell and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–15 and ‘Genetic identification of S. marinoi’.
Supplementary Tables
Supplementary Tables 1–5.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hattich, G.S.I., Jokinen, S., Sildever, S. et al. Temperature optima of a natural diatom population increases as global warming proceeds. Nat. Clim. Chang. (2024). https://doi.org/10.1038/s41558-024-01981-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41558-024-01981-9