Abstract
To efficiently combat viral infectious diseases, it is important to develop broadly applicable countermeasures, and efficient antiviral systems can be developed by elaborating the relationship of antiviral efficiency with the interactions between antiviral agents and viruses. In the present study, conjugated polymer (CP)-based photodynamic therapy was used to inhibit RNA virus infections. A severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pseudotyped virus composed of an SARS-CoV-2 envelope coated with the S protein and luciferase RNA genome was employed to assess antiviral efficiency. Three cationic CPs with different backbone structures, fluorene-co-phenylene (PFP), thiophene (PMNT), and phenylene vinylene (PPV), exhibit different photoinactivation effects. The highly efficient photoinactivation of PPV and PMNT is derived from the complete photodegradation of spike proteins, nucleocapsid proteins and nucleic acids of SARS-CoV-2 after binding to the viral spike proteins. Although PFP showed the highest efficiency in the photodegradation of spike proteins due to its strong binding affinity, ineffective viral inhibition was observed, which occurred because the viral gene was partially damaged under light irradiation and the process of delivering the viral gene to cells received assistance. This work preliminarily reveals the effect of CP-virus interactions on their photoinactivation activity and should be beneficial for further research on the development of highly efficient antiviral PDT agents.
Similar content being viewed by others
Introduction
Historically, viral epidemics and pandemics, such as SARS-CoV (2003), H1N1 (2009), and MERS-CoV (2012), have posed significant threats to human health1,2,3. At the end of 2019, the COVID-19 pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) rapidly wreaked havoc worldwide4. Addressing future outbreaks of emerging viruses has become a persistent challenge for humans. Vaccination, as the best preventive tool, has become widely used in the fight against viruses5. However, during a public health emergency, suitable vaccines are usually lacking because vaccines are specific to viruses and extensive time and resources are necessary to develop vaccines6. Moreover, other viruses, such as human immunodeficiency virus (HIV) and influenza viruses, exhibit a high genetic variability and cannot be controlled by vaccines7,8. As another important tool, antiviral drugs inhibit viral infection by affecting critical aspects of the viral life cycle9,10,11. Although these drugs are highly effective in treating certain viruses, their clinical application is limited due to drug resistance, which originates from the high mutation rate in viruses, and harsh side effects12,13. Therefore, it is of paramount importance to develop countermeasures that are generally applicable to combat viral diseases.
Photodynamic therapy (PDT) has shown a powerful ability to address the issues encountered with vaccines and antiviral drugs. PDT acts independently of the host response or viral replication processes and directly inactivates viruses by disrupting viral structures (nucleic acids, proteins and lipids) through reactive oxygen species (ROS) generated from light-irradiated photosensitizers14,15. Due to the physical damage of viral structures, the development of viral resistance is unlikely, and PDT treatment does not depend on the specific interaction with receptors; thus, PDT exhibits broad-spectrum antiviral activities16,17. In view of these advantages, PDT will become an alternative modality for treating viral diseases. Conjugated polymers (CPs) exhibit a strong light-harvesting ability and good ability to generate ROS; thus, CPs have been established as efficient photosensitizers against bacteria, fungi and biofilms18,19,20,21,22,23,24,25,26,27. In our previous work, cationic CPs could bind with proteins through electrostatic interactions and efficiently photoinactivated the proteins28; therefore, photodegrading the proteins of viruses to combat SARS-CoV-2 with cationic CPs should be feasible. SARS-CoV-2 is a novel β-coronavirus, and structurally, the virion is composed of an envelope and a helical capsid, in which the envelope contains membrane (M) proteins, envelope (E) proteins coated with spike (S) proteins, and nucleocapsid (N) proteins bound to the RNA genome to form the capsid29,30,31. Among these four structural proteins, the S protein is responsible for receptor binding and mediates membrane fusion, virus attachment and entry into host cells; thus, the S protein is the most important target in the development of anti-SARS-CoV therapeutics and vaccines32,33. The E protein is very important for virus infectivity34. The N protein of SARS-CoV-2 directly protects nucleic acids from the environment and participates in the transcription and replication of the viral genome35,36. The multiple functions of the N protein make it an important target of antiviral drugs37. Whitten’s research group demonstrated that cationic poly-phenylene ethynylene (CPE) and oligo-phenylene ethynylene (OPE) exhibited significant antiviral activity against two nonenveloped viruses (T4 and MS2 bacteriophages) under light irradiation38. Very recently, they found that OPEs inactivated SARS-CoV-2 with high efficiency under light irradiation39. However, the interaction modes between CPs and viruses were not clearly investigated and are directly related to their antiviral efficiency. Therefore, it is necessary to further investigate and uncover the interactions between CPs with viruses and their structural proteins to design efficient antiviral PDT systems.
Herein, the interactions between spike proteins, nucleocapsid proteins of SARS-CoV-2 and three cationic CPs (their chemical structures are shown in Fig. 1a) with different π-conjugated backbone structures (fluorene-co-phenylene: PFP, thiophene: PMNT, phenylene vinylene: PPV) were studied. Using SARS-CoV-2 pseudotyped viruses as a model, the photoinduced inactivation of RNA viruses by three cationic CPs was further explored. These three CPs were chosen for the following reasons: (1) The positive charges of CPs favor electrostatic binding to negatively charged virus proteins, and the π-conjugated backbone structures of CPs further enhance their binding affinities through hydrophobic interactions. The strong interaction between CPs and a virus is essential for photoinactivation of the virus. (2) The different π-conjugated backbone structures can endow three cationic CPs with different ROS generation abilities, and the flexibility of these three backbone structures influence their interaction with viruses, which affect their virus killing efficiency. Thus, this work aimed at providing a deep understanding of the relationship between the interactions of CPs and viruses with their photoinactivation efficiency.
Experimental section
Materials and methods
Cationic conjugated polymers (CPs)− PFP40, PMNT41, and PPV42 were synthesized according to the procedures in the literature. 2′,7′-Dichlorofluorescein diacetate (DCFH-DA) was purchased from Biyuntian (Shanghai, China). The S protein (S1 + S2, ECD), S protein (RBD) and N protein of SARS-CoV-2 used in the photoinduced degradation measurements were purchased from Genscript (Nanjing, China; catalog nos. Z03481-100 and Z03480-100). A schematic of the protein primary structures is shown in Figs. S1 and S2. High-performance precast mini polyacrylamide gels (SurePAGE gels) with homogeneous concentrations of 12%, broad multicolor prestained protein standard and tris-MOPS-SDS running buffer power were also purchased from Genscript (catalog nos. M00667, M00624-1250 and M00138). CBB Fast Staining solutions (catalog no. PA101) were obtained from TIANGEN biotech (Beijing) CO. LTD. The SARS-CoV-2 N gene coated with the capsid protein of MS2 phage was purchased from Zeesan Biotech (Xiamen, China). The PrimeScriptTM II 1st Strand cDNA Synthesis Kit was purchased from TaKaRa Bio Inc. Forward primer (N1) and reverse primer (N2) were purchased from Invitrogen Corporation. HotMaster Taq DNA polymerase and its corresponding buffer were purchased from TIANGEN Biotech. The pseudovirus incorporated with the SARS-CoV-2 S protein and the reporter gene firefly luciferase and HEK-293T cells overexpressing human ACE2 receptors were supplied by Sino Biological Inc., and the infection experiment was conducted in a biosafety level 2 (BSL-2) lab in Sino Biological Inc. (Beijing, China). Fetal bovine serum (FBS) was purchased from Sijiqing Biological Engineering Material (Hangzhou, China). Dulbecco’s modified Eagle’s medium (DMEM) was purchased from HyClone/Thermo Fisher (Beijing, China).
UV‒Vis absorption spectra were taken on a JASCO V-570 spectrophotometer. Fluorescence spectra were measured using a Hitachi F-4500 fluorometer equipped with a xenon lamp excitation source. Fluorescence and bioluminescence intensities were obtained on a microplate reader (BIO-TEK Synergy HT) using black 96-well plates (Thermo Scientific, lighttight, flat bottom, nonsterile). Isothermal titration microcalorimetry measurements were conducted on a TAM III (TA) instrument. Sodium dodecyl sulfate‒polyacrylamide gel electrophoresis (SDS‒PAGE) experiments were performed with a Mini-PROTEIN Tetra System from Bio-Rad. The images of the SDS‒PAGE gels and agarose gels were obtained with a Bio-Rad Molecular Imager ChemiDocXRS system.
Reactive oxygen species (ROS) measurements
The generation of ROS was probed by 2’,7’-dichlorofluorescin (DCFH), which converts to highly fluorescent 2,7-dichlorofluorescein (DCF) in the presence of ROS. The activated DCFH solutions were prepared by alkalization of 2,7-dichlorofluorescein diacetate (DCFH-DA). Then, 10 µL of the conjugated polymer solutions were added to 90 µL of the activated DCFH (40 µM) solutions (the final concentrations of the three conjugated polymers were all 20 µM). The mixed solutions were irradiated under white light (20 mW/cm2), and the fluorescence intensity of DCF at 524 nm (excited at 488 nm) was measured every minute.
Protein photodegradation measurements
The photodegradation effects of the SARS-CoV-2 S protein and N protein by the three CPs were investigated by SDS‒PAGE. A total of 300 μg/mL of S protein or 150 μg/mL of N protein were incubated with 50 μM of three CPs in the dark for 15 min at room temperature. Then, the mixture solutions were irradiated under white light (75 mW/cm2) for 2–10 min. The light-irradiated samples were denatured in 2× Tris-Tricine-SDS‒PAGE loading buffer at 100 °C for 5 min. After these treatments, the samples were loaded in a vertical SDS‒PAGE system. SDS‒PAGE was run on gels with homogeneous concentrations of 12% in Tris-MOPS-SDS running buffer (50 mM Tris Base, 50 mM MOPS, 1 mM EDTA, and 0.1% SDS) for 30 min at 200 V. The resultant gels were washed with dd-H2O and stained with Coomassie brilliant blue.
Interactions between CPs and SARS-CoV-2 proteins
The binding affinity between CPs and SARS-CoV-2 S protein or N protein was measured by isothermal titration microcalorimetry (ITC). The reference cell was filled with 765 µL of PBS buffer. The titration experiments consisted of injecting 10 µL (33 injections) of the 100 µM CP solution into the sample cells that were initially loaded with 600 µL of 40 μg/mL S protein or 20 μg/mL N protein solution. The injection interval was 10 min, and each injection lasted 20 s to ensure that the heat flow returned to baseline before the next injection. The system was stirred at 90 rpm with a gold propeller. The dilution heat of the CP solution was subtracted from the corresponding observed enthalpy curve of CP with the S protein or N protein solution. The binding constants (Ka) of CPs with these two proteins were obtained by fitting the ITC curves.
Pseudovirus-based photoinactivation assay
A pseudovirus incorporated with the SARS-CoV-2 S protein and the reporter gene firefly luciferase were used as the model virus, and HEK-293T/ACE2 cells were the host cells. A total of 1010 copies/mL pseudoviruses were incubated with different concentrations of CPs (PFP, PMNT and PPV) in the dark for 15 min and irradiated with white light (75 mW/cm2) for 30 min. Then, the mixture was added to HEK-293T/ACE2 cells in a 96-well plate. After incubation for 24 h, the virus-containing medium was replaced with fresh medium and cultured for another 24 h. Finally, pseudovirus infections were measured using the ONE-Glo™ Luciferase Assay System (Promega).
Gene photodegradation measurements
The SARS-CoV-2 N gene coated with the capsid protein of MS2 phage was used in the gene photodegradation measurements. A total of 1012 copies/mL of the N gene were incubated with 50 μM of three CPs in the dark for 15 min at room temperature, and then the mixed solutions were irradiated under white light (75 mW/cm2) for 5 min. After treatment with CPs and/or light irradiation, the gene samples were reverse transcribed and amplified by PCR in one step. The 25 μL reaction mixtures consisted of 2 μL of 108 copies/mL gene sample, 1 μL of 50 μM oligo dT primer, 2 μL of 10 mM dNTPs, 5 μL of 5×PrimeScript II Buffer, 0.5 μL of 40 U/μl RNase Inhibitor, 1 μL of 200 U/μl PrimeScript II RTase, 1 μL of 10 μM forward primer, 1 μL of 10 μM reverse primer, 0.5 μL of 2.5 U/μL HotMaster Taq DNA polymerase, and 2.5 μL of 10×HotMaster Taq Buffer. The reverse transcription and PCR conditions were programmed as follows: 30 min at 42 °C for reverse transcription, 2 min at 95 °C for predenaturation, then 30 cycles of 20 s at 95 °C, 20 s at 55 °C, and 40 s at 65 °C, and 5 min at 65 °C for final extension. The amplified samples were loaded onto a freshly prepared agarose gel and used for the electrophoresis assay.
Results and discussion
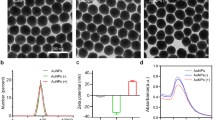
The photophysical properties and ROS generation abilities of CPs in aqueous solutions were measured, and as shown in Fig. 1b, the maximum absorption wavelengths of PFP, PMNT and PPV are 390 nm, 395 nm, and 448 nm, respectively. The emission peaks are at 433 nm, 532 nm, and 557 nm. These spectral data are summarized in Table S1. All three CPs display strong adsorption in both the near ultraviolet and visible regions, and spectral overlaps between the absorption of CPs and the spectral power distribution of the white light source are shown in Fig. S3; this finding guarantees that white light can be used for the subsequent photoinactivation of proteins and viruses. An ROS-sensitive probe (2′,7′-dichlorofluorescein diacetate, DCFH-DA) was utilized to characterize the generation of ROS from the three CPs (Fig. 1c). In the presence of ROS, activated 2′,7′-dichlorofluorescein (DCFH) by alkalization of DCFH-DA can be converted to the highly fluorescent 2′,7′-dichlorofluorescein (DCF). The fluorescence intensities of DCF treated with PFP and PPV increase dramatically and reach a plateau after being irradiated with white light for 3 min, while the fluorescence intensity of DCF treated with PMNT increases slowly. These results confirm that all three CPs can sensitize the surrounding oxygen to generate ROS under irradiation with white light and that the π-conjugated backbone structures endow the CPs with different ROS generation abilities, which are in the following order: PPV > PFP > PMNT.
We first tested the photoinactivation of S proteins of SARS-CoV-2 and their interactions with three CPs. The photoinactivation of the S protein by PFP, PMNT and PPV was performed as illustrated in Fig. 2a, and the damage to the S protein was analyzed with SDS polyacrylamide gel electrophoresis (SDS PAGE). Here, two lengths of S proteins were selected to study the photoinduced degradation of protein after treatment with three CPs. One is the full-length S protein, which plays an important role in the infectious process from receptor binding to viral entry into host cells. The other is the S protein segment that contains the RBD domain, which is mainly responsible for the binding between the virus and receptor in host cells, and the segment is denoted as the S protein (RBD). The photoinactivation results are shown in Fig. 2b, Fig. S4 and Fig. S5a. The bands of S proteins exposed to white light alone or treated with three CPs in the dark exhibited little change from the band of S proteins without any treatment. In contrast, the bands of S proteins treated by three CPs under white light irradiation displayed a significant decrease, indicating that S proteins were damaged by the ROS sensitized by the irradiation of CPs. As shown in Fig. 2c, the quantity of S protein treated by PFP, PMNT and PPV under white light irradiation was reduced to 2.0%, 37.3% and 18.2%, respectively. Similarly, the quantity of S protein (RBD) was obviously decreased after treatment with the three CPs and light irradiation. The photoinactivation efficiency of the S protein (RBD) decreased in the following order: PFP > PPV > PMNT (Fig. S5b). These results indicate that all three CPs can efficiently inactivate the S protein through photodegradation of the RBD region and the other functional domains.
a Schematic of the photodegradation assays. S proteins were incubated with CPs for 15 min and then irradiated by white light for 10 min. The light-irradiated mixtures were analyzed by SDS‒PAGE. b Gel picture and (c) Quantitative analysis. d ITC analysis of the binding affinity between CPs and the S protein.
The PFP with a moderate ROS generation ability exhibited the best performance in the photodegradation of S proteins, which might be attributed to the affinity difference between the CPs and S proteins. Full-length S protein was used to study its interaction with three CPs. The binding affinities between the three CPs and the S protein were investigated by isothermal titration microcalorimetry (ITC) measurements. As presented in Fig. 2d, the observed enthalpy changes (ΔHobs) were initially endothermic, then became less positive, and finally approached zero for PFP and PPV. The values of ΔHobs changed from exothermic to zero for PMNT. The binding constants (Ka) and enthalpy change (ΔH) were obtained by fitting the ITC curves using the binding model of the single set of identical sites, together with Gibbs free energy change (ΔG) and entropy change (ΔS), as shown in Fig. S6a. The Ka value of PFP was 17.4×106 M−1, one order of magnitude higher than that of the other two CPs, which suggested that the higher photodegradation efficiency of PFP to S protein presumably resulted from the strong affinity between the CPs and S protein. All derived binding constants Ka were higher than 106 M−1, and the ΔS values were positive, confirming the hydrophobic interactions with the S protein. In this context, ΔH values were positive for PFP and PPV with the S protein, while they were negative for PMNT, suggesting that stronger electrostatic interactions were observed for PFP and PPV binding with the S protein. The stronger electrostatic attraction was generally accompanied by dehydration, which was an endothermic process. In addition, the relatively strong steric hindrance due to the rigid backbone structures of PFP and PPV made the electrostatic binding of PFP and PPV to the S protein an endothermic process. The zeta potential (ζ) and fluorescence spectra provided direct evidence for the binding of CPs to the S protein. The ζ for the S protein was −11 mV and became more positive after the protein-bound with CPs (Fig. S6b), indicating that an electrostatic attraction occurs between CPs and the S protein. With the addition of the S protein, the fluorescence intensity of CPs increased by 11~23% (Fig. S7). This may occur because π − π stacking in the conjugated backbone of CPs is prevented when cationic CPs wrap around the negatively charged protein through electrostatic and hydrophobic interactions.
We further investigated the photodegradation of the N protein of SARS-CoV-2 by three CPs (Fig. 3a). As shown in Fig. 3b, the bands of N protein treated by PFP, PPV and PMNT showed a distinct decrease only after 2 min of light irradiation. No other bands were observed in the whole lane in SDS PAGE (Fig. S8), which indicated that the reduced proteins were completely damaged by the ROS sensitized by conjugated polymers. The quantitative analysis of the SDS PAGE results is shown in Fig. 3c. The inactivation efficiency of the N protein under light irradiation decreased in the following order: PMNT (62.0%) > PPV (53.7%) > PFP (42.7%). In this context, PMNT with relatively low ROS generation ability surpassed the performance of PPV and PFP in the photodegradation of N protein. The binding affinities between the three CPs and the N protein were investigated by ITC measurements, and similar interaction modes were obtained for the N protein with CPs, as for the S protein (Fig. 3d). Ka values were obtained by fitting the ITC curves, and the largest Ka value (4.51 × 106 M−1) ensured that PMNT attained the highest photodegradation efficiency toward N protein. Since the Ka value of PPV binding to the N protein (0.91 × 106 M−1) was close to that of PFP binding to the N protein (1.58 × 106 M−1), the photodegradation efficiency was also determined by their ability to generate ROS. The zeta potential (ζ) for the N protein of SARS-CoV-2 was −21 mV and became more positive after binding with CPs, which confirmed the electrostatic attractions between the N protein and CPs (Fig. S9). The addition of N protein to CP solution induces an obvious decrease in the fluorescence intensity of CPs (Fig. S10). Considering that the N protein (~50 kDa) is much smaller than the S protein (~135 kDa), the binding of the N protein to CPs cannot prevent the π − π stacking of CPs. Their binding can only decrease the electrostatic repulsion between the side chains of CPs, which in turn promotes interchain π − π stacking and leads to a decrease in the fluorescence intensity of CPs. The photodegradation of the S and N proteins of SARS-CoV-2 with PFP, PPV and PMNT and their corresponding binding constants are summarized in Table 1. From these results, it can be concluded that the photodegradation efficiency of proteins is mainly determined by the affinity between CP and proteins and then by their ability to generate ROS.
a Schematic of the photodegradation assays. N proteins were incubated with CPs for 15 min and then irradiated by white light for 2 min. The light-irradiated mixtures were analyzed by SDS‒PAGE. b Gel picture and (c) statistical analysis. d ITC analysis of the binding affinity between CPs and the N protein.
The photoinactivation effects on SARS-CoV-2 pseudoviruses were subsequently evaluated, as illustrated in Fig. 4a. The pseudoviruses were composed of the SARS-CoV-2 envelope coated with the S protein and RNA genome with luciferase as a reporter gene. In this assay, the infected cells were HEK293T cells overexpressing angiotensin-converting enzyme 2 receptors (HEK293T/ACE2)43,44. SARS-CoV-2 pseudoviruses recognize and bind to host receptors, and subsequently, the viral envelope fuses with the host cell membrane, completing the infection process. Thus, the infections were analyzed by the luciferase activities. The binding of CPs with SARS-CoV-2 pseudoviruses exhibited a slight influence on virus entry mediated by the ACE2 receptor, and the virus remained infectious for host cells, as expected. After light irradiation, different photoinactivation effects were observed for the three conjugated polymers. PMNT and PPV showed dose-dependent photoinactivation of pseudovirus, while PFP exhibited little effect on the photoinactivation of pseudovirus at the studied concentrations, as shown in Fig. 4b–d. For PFP-treated pseudoviruses, the values of relative light unit (RLU) decreased, but the inhibition rate only increased from 32% to 38% with increasing PFP concentration to 50 μM (Fig. 4e, the blue line). For PMNT- and PPV-treated pseudoviruses, the RLU values decreased by three orders of magnitude when the concentrations increased to 50 μM for PMNT and 5 μM for PPV (Fig. 4e). These results indicated that compared to PMNT, PPV was more effective in photoinactivating pseudoviruses; thus, the antiviral activity was specific. Interestingly, an increase in the RLU values of the infected cells was observed when pseudoviruses were treated with 50 μM PFP under dark conditions (Fig. 4b). In contrast, PMNT-treated pseudovirus showed no influence on the RLU values (Fig. 4c), and PPV-treated pseudovirus caused a decrease in the RLU value with increasing PPV concentration (Fig. 4d). These results suggested that PFP functioned as a vector to assist SARS-CoV-2 N gene delivery and transfection into HEK293T/ACE2 cells. Therefore, through the combination of low gene photodegradation efficiency and gene delivery capability, the photoinactivation of pseudovirus by PFP is ineffective45.
a Schematic of the photoinactivation assays. A total of 1010 copies/mL pseudoviruses bearing S protein were incubated with different concentrations of CPs for 15 min and then irradiated with white light for 30 min. The light-irradiated mixtures were used to infect HEK293T/ACE2 cells, and the infections were analyzed by measuring luciferase activity. Infection of HEK293T/ACE2 cells with (b) PFP-, (c) PMNT-, and (d) PPV-treated pseudovirus. e Variation in pseudovirus inhibition with CP concentrations.
The different photoinhibition rates might be related to efficiency by which PPV and PMNT perform photodegradation of genes; therefore, the photodegradation of the SARS-CoV-2 N gene coated with the capsid protein from the MS2 phage by CPs was performed, and the results are presented in Fig. 5. Based on the results from agarose gel electrophoresis analysis, PPV was most efficient in degrading the SARS-CoV-2 N gene, while PFP presented the lowest photodegradation efficiency under identical conditions. As discussed above, the three CPs with different backbone structures exhibited different photoinactivation efficiencies against SARS-CoV-2 pseudovirions. PPV and PMNT bound to the virus surface through strong binding ability with the S protein, which showed a slight influence on virus entry mediated by the ACE2 receptor and maintained the ability of the virus to infect host cells. When the mixtures of PPV/virus and PMNT/virus were light irradiated, the ROS sensitized by PPV or PMNT caused both proteins and genes to completely cleave, which prevented SARS-CoV-2 from attaching to the ACE2 receptor and damaged generic materials of viruses, leading to effective photoinactivation of viruses. In comparison with that of PPV and PMNT, virus infection was enhanced after binding with PFP due to the gene delivery capability of PFP. Light irradiation mainly caused the protein structures to be disrupted, and the remaining gene could be efficiently delivered into cells using PFP as a vector, resulting in failure to effectively photoinactivate viruses.
a Schematic illustration of the photodegradation assays. b Gel electrophoresis image. A total of 1012 copies/mL pseudoviruses incorporating the SARS-CoV-2 N gene were incubated with 50 μM CPs for 15 min and then irradiated with white light (75 mW/cm2) for 5 min. The gene number of light-irradiated samples was amplified by one round of PCR and then analyzed by agarose gel electrophoresis.
Conclusion
In summary, we have demonstrated that three cationic CPs with different backbone structures exhibit different photoinactivation effects against RNA viruses. PPV and PMNT cause viruses to be effectively inactivated under light irradiation, while SARS-CoV-2 pseudotyped viruses remain infectious after treatment with PFP, which was determined by the interactions between CPs with the proteins and genes of viruses. All three CPs can photodegrade the proteins of SARS-CoV-2 efficiently, which prevents viral attachment to the ACE2 receptor, and higher photodegradation efficiency was observed for the CPs with stronger binding affinity to proteins. PPV can also completely photodegrade nucleic acids after protein degradation; thus, PPV exhibited the highest efficiency in photoinactivation of viruses. PFP only causes the viral gene to be partially damaged under light irradiation and combines with the gene delivery ability, leading to the ineffective photoinactivation of viruses. Our findings provide a fundamental understanding of the relationships between CP structures, CP-virus interactions and photoinactivation activity, which would be beneficial for developing highly efficient PDT agents against SARS-CoV-2 as well as other viruses threatening human health.
References
Corman, V. M., Muth, D., Niemeyer, D. & Drosten, C. Hosts and sources of endemic human coronaviruses. Adv. Virus Res. 100, 163–188 (2018).
Liu, J. et al. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoC, and 2019-nCoV. J. Med. Virol. 92, 491–494 (2020).
Neumann, G., Noda, T. & Kawaoka, Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459, 931–939 (2009).
Wang, C., Horby, P. W., Hayden, F. G. & Gao, G. F. A novel coronavirus outbreak of global health concern. Lancet 395, 470–473 (2020).
Ferguson, N. M. et al. Strategies for mitigating an influenza pandemic. Nature 442, 448–452 (2006).
Pardi, N. & Weissman, D. Development of vaccines and antivirals for combating viral pandemics. Nat. Biomed. Eng. 4, 1128–1133 (2020).
Leslie, A. J. et al. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10, 282–289 (2004).
Nelson, M. I. & Holmes, E. C. The evolution of epidemic influenza. Nat. Rev. Genet. 8, 196–205 (2007).
Ahidjo, B. A., Loe, M. W. C., Ng, Y. L., Mok, C. K. & Chu, J. J. H. Current perspective of antiviral strategies against Covid-19. ACS Infect. Dis. 6, 1624–1634 (2020).
Geraghty, R. J., Aliota, M. T. & Bonnac, L. F. Broad-spectrum antiviral strategies and nucleoside analogues. Viruses 13, 667 (2021).
De Clercq, E. & Li, G. Approved antiviral drugs over the past 50 years. Clin. Microbiol. Rev. 29, 695–747 (2016).
Calmy, A., Hirschel, B., Cooper, D. A. & Carr, A. Clinical update: Adverse effects of antiretroviral therapy. Lancet 370, 12–14 (2007).
Zhou, J., Krishnan, N., Jiang, Y., Fang, R. H. & Zhang, L. Nanotechnology for virus treatment. Nano Today 36, 101031 (2021).
Sobotta, L., Skupin-Mrugalska, P., Mielcarek, J., Goslinski, T. & Balzarini, J. Photosensitizers mediated photodynamic inactivation against virus particles. Mini-Rev. Med. Chem. 15, 503–521 (2015).
Costa, L., Faustino, M. A., Neves, M. G., Cunha, A. & Almeida, A. Photodynamic inactivation of mammalian viruses and bacteriophages. Viruses 4, 1034–1074 (2012).
Yin, H. et al. Photoinactivation of cell-free human immunodeficiency virus by hematoporphyrin monomethyl ether. Lasers Med. Sci. 27, 943–950 (2012).
Wiehe, A., O’Brien, J. M. & Senge, M. O. Trends and targets in antiviral phototherapy. Photochem. Photobiol. Sci. 18, 2565–2612 (2019).
Lu, L. D. et al. Biocidal activity of a light-absorbing fluorescent conjugated polyelectrolyte. Langmuir 21, 10154–10159 (2005).
Wang, Y. et al. Understanding the dark and light-enhanced bactericidal action of cationic conjugated polyelectrolytes and oligomers. Langmuir 29, 781–792 (2013).
Parthasarathy, A. et al. Conjugated polyelectrolytes with imidazolium solubilizing groups. Properties and application to photodynamic inactivation of bacteria. ACS Appl. Mater. Interfaces 7, 28027–28034 (2015).
Pappas, H. C. et al. Antifungal properties of cationic phenylene ethynylenes and their impact on β-glucan exposure. Antimicrob. Agents Chemother. 60, 4519–4529 (2016).
Xing, C. et al. Conjugated polymers for light-activated antifungal activity. Small 8, 524–529 (2012).
Bai, H. et al. A supramolecular antibiotic switch for antibacterial. Regul. Angew. Chem. Int. Ed. 54, 13208–13213 (2015).
Wang, B., Wang, M., Mikhailovsky, A., Wang, S. & Bazan, G. C. A membrane-intercalating conjugated oligoelectrolyte with high-efficiency photodynamic antimicrobial activity. Angew. Chem. Int. Ed. 56, 5031–5034 (2017).
Wu, W. et al. A cross-linked conjugated polymer photosensitizer enables efficient sunlight-induced photooxidation. Angew. Chem. Int. Ed. 58, 3062–3066 (2019).
Jagadesan, P. et al. Light-activated antifungal properties of Imidazolium- functionalized cationic conjugated polymers. Chem. Mater. 32, 6186–6196 (2020).
Zhang, P. et al. Biofilm inhibition and elimination regulated by cationic conjugated polymers. ACS Appl. Mater. Interfaces 9, 16933–16938 (2017).
Duan, X., Liu, L., Feng, X. & Wang, S. Assemblies of conjugated polyelectrolytes with proteins for controlled protein photoinactivation. Adv. Mater. 22, 1602–1606 (2010).
Wu, F. et al. A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020).
Chan, J. F.-W. et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 9, 221–236 (2020).
Chen, Y., Liu, Q. & Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 92, 418–423 (2020).
Du, L. et al. The Spike Protein of SARS-CoV - a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 7, 226–236 (2009).
Samrat, S. K., Tharappel, A. M., Li, Z. & Li, H. Prospect of SARS-CoV-2 Spike Protein: Potential role in vaccine and therapeutic development. Virus Res. 288, 198141 (2020).
Oberfeld, B. et al. SnapShot: COVID-19. Cell 181, 954 (2020).
Peng, T.-Y., Lee, K.-R. & Tarn, W.-Y. Phosphorylation of the Arginine/Serine Dipeptide-rich motif of the severe acute respiratory syndrome coronavirus nucleocapsid protein modulates its multimerization, translation inhibitory activity and cellular localization. FEBS J. 275, 4152–4163 (2008).
Terry, J. S. et al. Development of a SARS-CoV-2 nucleocapsid specific monoclonal antibody. Virology 558, 28–37 (2021).
Kang, S. et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B 10, 1228–1238 (2020).
Wang, Y. et al. Cationic Phenylene Ethynylene polymers and oligomers exhibit efficient antiviral activity. ACS Appl. Mater. Interfaces 3, 2209–2214 (2011).
Monge, F. A. et al. Highly effective Inactivation of SARS-CoV-2 by conjugated polymers and oligomers. ACS Appl. Mater. Interfaces 12, 55688–55695 (2020).
Liu, B., Wang, S., Bazan, G. C. & Mikhailovsky, A. Shape-adaptable water-soluble conjugated polymers. J. Am. Chem. Soc. 125, 13306–13307 (2003).
Ho, H.-A. et al. Colorimetric and Fluorometric detection of nucleic acids using cationic polythiophene derivatives. Angew. Chem. Int. Ed. 41, 1548–1551 (2002).
Yuan, H. et al. Cationic conjugated polymers for discrimination of microbial pathogens. Adv. Mater. 26, 4333–4338 (2014).
Ou, X. et al. Characterization of Spike Glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 11, 1620 (2020).
Nie, J. et al. Quantification of SARS-CoV-2 neutralizing antibody by a pseudotyped virus-based assay. Nat. Protoc. 15, 3699–3715 (2020).
Feng, X. et al. A highly emissive conjugated polyelectrolyte vector for gene delivery and transfection. Adv. Mater. 24, 5428–5432 (2012).
Acknowledgements
We are grateful for financial support from the National Key R&D Program of China (2018YFE0200700), National Natural Science Foundation of China (22021002, 22020102005, and 22022705), and the CAS-Croucher Funding Scheme for Joint Laboratories.
Author information
Authors and Affiliations
Contributions
R.Q. and F.L. performed all experiments and authored the paper. Y.Z. and Q.S. contributed to the PCR characterizations and discussion. Y.H., H.B., and L.L. guided the synthesis of the three polymers. S.W. conceived and directed the whole study and writing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Qi, R., Lv, F., Zeng, Y. et al. Photoactive conjugated polymer-based strategy to effectively inactivate RNA viruses. NPG Asia Mater 15, 17 (2023). https://doi.org/10.1038/s41427-022-00455-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41427-022-00455-8
This article is cited by
-
aPDI meets PPE: photochemical decontamination in healthcare using methylene blue—where are we now, where will we go?
Photochemical & Photobiological Sciences (2024)