Abstract
The centrosome assembles a bipolar spindle for faithful chromosome segregation during mitosis. To prevent the inheritance of DNA damage, the DNA damage response (DDR) triggers programmed spindle multipolarity and concomitant death in mitosis through a poorly understood mechanism. We identified hornerin, which forms a complex with checkpoint kinase 1 (Chk1) and polo-like kinase 1 (Plk1) to mediate phosphorylation at the polo-box domain (PBD) of Plk1, as the link between the DDR and death in mitosis. We demonstrate that hornerin mediates DDR-induced precocious centriole disengagement through a dichotomous mechanism that includes sequestration of Sgo1 and Plk1 in the cytoplasm through phosphorylation of the PBD in Plk1 by Chk1. Phosphorylation of the PBD in Plk1 abolishes the interaction with Sgo1 and phosphorylation-dependent Sgo1 translocation to the centrosome, leading to precocious centriole disengagement and spindle multipolarity. Mechanistically, hornerin traps phosphorylated Plk1 in the cytoplasm. Furthermore, PBD phosphorylation inactivates Plk1 and disrupts Cep192::Aurora A::Plk1 complex translocation to the centrosome and concurrent centrosome maturation. Remarkably, hornerin depletion leads to chemoresistance against DNA damaging agents by attenuating DDR-induced death in mitosis. These results reveal how the DDR eradicates mitotic cells harboring DNA damage to ensure genome integrity during cell division.
Similar content being viewed by others
Introduction
The DNA damage response (DDR) transmits signals to cell cycle checkpoints and coordinates DNA repair and apoptosis to prevent the generation of deleterious mutations and carcinogenesis [1, 2]. Of note, the core DNA repair machinery is suppressed during mitosis due to the fusogenic potential of mitotic telomeres [3]. Despite the effectiveness of cell cycle checkpoints in safeguarding genomic integrity, excessive DNA damage or weakened G2/M checkpoint probably results in premature or inappropriate onset of mitosis and concomitant mitotic derangement, which are resolved by delayed mitosis-associated cell death, mitotic catastrophe, during mitosis or following G1 [4,5,6]. Although the DDR produces binuclear cells through the ATR-Chk1-Aurora B pathway and concomitant cell death or senescence in the next G1 phase [7], the intricate mechanism of DDR-mediated death in mitosis (DiM), which is an indispensable pathway in mitotic catastrophe, is largely unknown.
The fundamental function of the centrosome, which comprises two centrioles in orthogonal configuration and surrounding pericentriolar material (PCM), is to nucleate and organize microtubules for cell migration, intracellular payload trafficking, and cell polarity in interphase and for the assembly of the mitotic spindle and the segregation of sister chromatids in mitosis [8]. The centriole is replicated once a cell cycle with template-dependent duplication, of which the cycle is remarkably similar to DNA replication, in S phase [9]. The first step of centriole duplication is the assembly of Plk4, STIL, and SAS-6 on the wall of the mother centriole [10]. Duplicated centriole pairs are separated in G2 to establish bipolar spindles by dissolving linker fibers between parental centrioles and disengaged at their proximal ends of parental centrioles by separase-mediated proteolysis of cohesin during anaphase after chromosome separation in an analogous manner. Similar to the protection of cohesin removal by Shugoshin 1 (Sgo1) and phosphatase PP2A at the centromeres in prophase, Sgo1 and a shorter Sgo1 splice variant (sSgo1) also safeguard centriole cohesion from the Plk1-mediated prophase pathway [11, 12]. In this regard, overexpression of separase or depletion of Sgo1 results in unscheduled chromosome separation and precocious centriole disengagement [13]. While supernumerary centrosomes caused by centrosome amplification (CA) through the dysregulation of the Plk4-STIL-SAS-6 module or precocious centriole disengagement are a conspicuous feature of chromosomal instability (CIN) cancers [14, 15], tumor cells undergo multipolar cell divisions by reconfiguring a multipolar spindle into a bipolar spindle through a centrosomal clustering mechanism [16, 17]. Although several DNA-damaging agents (DDAs) reportedly cause multipolar spindles [18], it is unclear how DDR triggers centrosome-mediated DiM during cell cycle progression.
During mitosis, the spindle assembly checkpoint (SAC) monitors kinetochore-microtubule attachment and spindle biorientation for faithful sister-chromatid segregation to two daughter cells [19]. In the presence of unattached or improperly attached kinetochores, the SAC delays anaphase onset via inhibition of the anaphase-promoting complex/cyclosome which activates separase through degradation of securin to cleave cohesin and, therefore, initiates sister-chromatid segregation [20]. Thus far, DDR proteins, such as ATM, ATR, Chk1, Chk2, BRCA2, MDC1, and 53BP1, have been reported to be involved in mitotic processes under unperturbed conditions [21,22,23,24] and DDR crosstalk with the SAC in DNA-damage-induced mitotic arrest [25]. For example, Chk1 phosphorylates Aurora B to delay anaphase onset in the presence of misattached kinetochores for faithful chromosome segregation by promoting BubR1 and Mps1 localization to kinetochores and to correct merotelic attachment by MCAK. Here, we report a molecular link, hornerin, that transmits signals from the DDR to the mitotic centrosome by mediating Plk1 phosphorylation by Chk1 and triggers DiM via precocious centriole disengagement and concomitant spindle multipolarity.
Results
DDR induces DiM via spindle multipolarity
To dissect DDR-induced mitotic catastrophe, we performed live-cell imaging with HeLa cells stably expressing GFP-tubulin after treatment with the mild DDA, benzo[a]pyrene (B[a]P), for 48 h to enrich the mitotic population and found that 70% of cells undergoing DiM harbored multipolar spindles (Fig. 1A, B, Fig. S1, and Movies 1–7). The DDA also produced polylobed progeny through regression in multipolar anaphase (Fig. 1C). DiM with a multipolar spindle configuration is thus a major pathway in mitotic catastrophe because most genotoxic agents drive the formation of multipolar mitotic spindles through the ATR-Chk1 pathway in the DDR (Fig. 1D, E, F, G), as previously described [26, 27]. Because B[a]P is known as a ligand for the aryl hydrocarbon receptor (AhR) and aggravates its own toxicity as a DDA by enhancing metabolic enzymes including CYP1A1, 1A2, and 1B1 [28], depletion of AhR expectedly decreased phospho-H2AX positive mitotic cells under B[a]P treatment and concurrently rescued B[a]P-induced multipolarity (Fig. S2). In contrast, etoposide-induced DNA damage and concomitant multipolarity were not rescued by AhR depletion, indicating that AhR is not directly involved in DDR-induced multipolarity. Although several abnormal conditions, such as centriole overduplication, precocious centriole disengagement, and pericentriolar material (PCM) fragmentation, clearly result in CA and the formation of multipolar spindles [18, 29], the mechanism underlying DDR-induced spindle multipolarity is not yet clear. Consistent with previous reports [30], DDAs caused centriole overduplication in G2 and ultimately generated CA during mitosis (Fig. 2A, B). Given that Plk4, STIL, and SAS-6 are critical for the initiation of centriole duplication [10], we examined protein levels after treatment with DDAs and found substantial increases in these three proteins (Fig. 2C). In stark contrast to spindle multipolarity, the increases in Plk4, STIL, and SAS-6 and DNA damage-induced centriole overduplication were not suppressed by any of the inhibitors of the DDR (Fig. 2C, D), indicating that the DDR induces centriole overduplication in an ATM/ATR-independent manner. Because the radiomimetic drug, doxorubicin, and replication stress reportedly cause precocious centriole disengagement in G2 and mitosis [31,32,33], we examined centriole configuration by time-lapse imaging of HeLa cells stably expressing GFP-centrin after treatment with DDAs. As expected, B[a]P caused premature centriole disengagement in G2, which in turn resulted in the formation of multipolar spindles during mitosis (Fig. 3A, B, C, D, and Movies 8, 9). Intriguingly, inhibition of ATR or Chk1 substantially rescued DNA damage-induced premature centriole disengagement (Fig. 3E), strongly suggesting that the DDR triggers multipolar spindle and concomitant DiM via precocious centriole disengagement rather than centriole overduplication. Similar perturbation of centrosome and centriole integrity by DDAs was observed in untransformed RPE-1 cells (Fig. 3F, G, and Fig. S3).
A–C HeLa cells treated with 0.5 µM B[a]P for 48 h were analyzed by live cell imaging for 10 h (A, n = 1609 cells). DiM (B, n = 356) and the fate of cells with multipolar spindles (C, n = 257) were analyzed. D, E After treatment with the indicated genotoxic stresses, HeLa cells were stained with the indicated antibodies, and the number of prometaphase cells with multipolar spindles was determined from 300 prometaphase cells in three independent experiments. F, G Twenty-four hours after treatment with 2 µM NU6027 as an ATR inhibitor (ATRi), 20 nM KU55933 as an ATM inhibitor (ATMi), 300 nM UCN-01 as a Chk1 inhibitor (Chk1i), 1 µM BML-277 as a Chk2 inhibitor (Chk2i), or 0.5 µM KU57788 as a DNA-PK inhibitor (DNA-PKi), HeLa cells were treated with 0.5 µM B[a]P for 48 h or 0.5 µM etoposide for 24 h, and multipolar spindles were analyzed (n = 400 centrioles from three independent experiments). Scale bars, 5 µm. Error bars, SEM. *p < 0.01 (two-tailed t test).
A, B After treatment with the indicated chemicals, HeLa cells were stained with the indicated antibodies and the number of centrioles in prometaphase cells was analyzed and plotted (n = 300 cells). C, D Twenty-four hours after treatment with the indicated inhibitors, HeLa cells were treated with the indicated chemicals and analyzed by immunoblotting and immunostaining (D, n = 300). Images of uncropped blots are provided as a Supplementary Material file. Scale bars, 5 µm. Error bars, SEM. *p < 0.01 (two-tailed t test).
A HeLa cells expressing GFP-centrin 2 were treated with B[a]P and analyzed by live cell imaging. Arrows point to disengaged centrioles. B–E Disengagement of centrioles in chemically treated HeLa cells was stained with the indicated antibodies and analyzed (n = 300). Ninein served as a marker for the mother centriole (B). F, G After treatment with the indicated DNA damaging agents, RPE-1 cells were stained with the indicated antibodies (F). The number of RPE-1 cells with centriole overduplication or premature centriole disengagement was analyzed as in C and D (G; n = 300 cells). Scale bars, 5 µm. Error bars, SEM. *p < 0.01 (two-tailed t test).
DDR interferes with the targeting of Plk1 and Sgo1 to the centrosome
We speculated that separase and Plk1 could be relevant targets of the DDR in DiM because both are best known for their roles in relieving sister chromatid and centriolar cohesion during the metaphase-anaphase transition and are associated with precocious centriole disengagement in perturbed conditions [11, 34]. DNA damage-induced precocious centriole disengagement was not reversed by the depletion of separase with siRNA targeting the 3’ noncoding sequence of the endogenous separase gene (Fig. 4A, B). However, in addition to inhibition of Plk1 activity [35], immunofluorescence microscopy showed that the localization of Plk1 in the centrosome substantially decreased in a Chk1-dependent manner after treatment with DDAs (Fig. 4C). Given that Plk1 phosphorylates not only the SA/Scc3 cohesin subunit to trigger cohesin dissociation from the chromosome arm and centriole without proteolytic activity of separase in the prophase pathway but also Sgo1 to target it to the centrosome and protect centriole cohesion during early mitosis [12, 36,37,38,39], we sought to investigate the relationship between Plk1 and Sgo1 in DDR-induced precocious centriole disengagement. Interestingly, the translocation of Sgo1 to the centrosome was drastically diminished in cells treated with the DDAs (Fig. 4D, E) and the interaction of Plk1 with Sgo1 was substantially decreased in an immunoprecipitation assay and an in vitro binding assay with cell lysates from cells treated with the DDAs (Fig. 4F, G). These data indicate that the DDR disrupts the interaction of Plk1 with Sgo1 to induce precocious centriole disengagement.
A, B Twenty-four hours after siRNA transfection, HeLa cells were transfected with plasmids containing WT or noncleavable (NC) mutant separase and treated with B[a]P for 48 h. The lysates were analyzed by Western blotting with the indicated antibodies. Prometaphase cells with centriole disengagement were counted, and the data were plotted (n = 300). C, D The intensity of Plk1 and Sgo1 in the centrosome after genotoxic stress treatment (n = 30). E Forty-eight hours after treatment with the indicated chemicals, centrosomes were purified and analyzed by Western blotting with the indicated antibodies. F Immunoprecipitates from chemically treated HeLa cells were analyzed by immunoblotting. G Lysates from plasmid transfected cells were incubated with Ni-beads immobilized with His-Sgo1. Pulled-down proteins were analyzed by immunoblotting. Images of uncropped blots and gels are provided as a Supplementary Material file. AU, arbitrary units, Scale bars, 5 µm. Error bars, SEM. *p < 0.01 (two-tailed t test).
Chk1 phosphorylates the polo-box domain of Plk1
To investigate the mechanism underlying the disruption of Plk1 translocation to the centrosome by Chk1, we sought to identify phosphorylation sites within the polo-box domain (PBD), which is responsible for both the substrate specificity and centrosomal localization of Plk1 [40,41,42], by immunoprecipitation with mass spectrometry (Fig. S4A). We identified Ser526, Ser529, and Thr539 in the PBD as candidate sites among 14 phosphorylation residues under B[a]P-treatment (Fig. S4B). As expected, changing three residues to phosphorylation-mimicking Asp (3D) abrogated the localization of Plk1 to the centrosome, whereas changing these residues to Ala (3A) did not affect targeting to the centrosome (Fig. 5A, B). We consistently found that the WT and 3 A mutant, but not the 3D mutant, successfully rescued spindle multipolarity under B[a]P-treatment (Fig. 5C). Next, we generated phospho-Ser526-specific, phospho-Ser529-specific, or phospho-Thr539-specific antibodies to confirm the phosphorylation of endogenous Plk1 and determine the corresponding kinase (Fig. S4C). Whereas three residues were phosphorylated in B[a]P-treated cells, Ser529 and Thr539 were phosphorylated mainly in Cdk1 inhibitor-treated cells arrested in the G2 phase and Ser526 in nocodazole-treated cells arrested in early mitosis (Fig. 5D). Consistent with this, phosphorylation of Ser529 and Thr539 decreased in cells released from G2 arrest (Fig. 5E). These data indicate that individual phosphorylation of Ser526, Ser529 and Thr539 is involved in cell cycle progression during G2 and mitosis. Similar to the 3D mutant, phosphorylated Plk1 also decreased at the centrosomes after treatment with DDA (Fig. S4D). Because the level of Plk1 at the centrosome was recovered by the Chk1 inhibitor (Fig. 4C), we tested whether Chk1 was the corresponding kinase for phosphorylation of the PBD in Plk1. Consistent with our findings, both ATR and Chk1 inhibitors abolished the phosphorylation of Ser529 and Thr539 induced by B[a]P treatment (Fig. 5F). In addition to phosphorylation of Thr210 during mitosis [43], Chk1 directly phosphorylated Ser529 and Thr539 in an in vitro kinase assay (Fig. 5G). To exclude the possibility of autophosphorylation by active Plk1, we confirmed the phosphorylation of a dominant-negative lysine 82 to arginine mutated Plk1 (K82R) [44] by Chk1 (Fig. 5H). Furthermore, mutation of Ser529 and Thr539 to Ala abrogated specific phosphorylation by Chk1 (Fig. 5G) and Chk1 directly interacted with Plk1 in an in vitro binding assay (Fig. 5I). These data indicate that Chk1 is a physiological kinase for phosphorylation at Ser529 and Thr539 in the PBD of Plk1 under DDR. However, the phosphorylation of Ser526 was not abolished by either ATR or Chk1 inhibitors and was not induced by Chk1 (Fig. 5F, G). Because the phosphorylation of Ser526 was substantially increased in nocodazole-treated cells arrested in early mitosis (Fig. 5D), we investigated whether mitotic kinases participate in this phosphorylation. Intriguingly, the Aurora A inhibitor, but not inhibitors against mitotic kinases including Cdk1, Plk1, and Aurora B, drastically abolished the phosphorylation of Ser526 (Fig. 5J). In addition to phosphorylation at Thr210 in late G2 and early mitosis [45], Aurora A strongly phosphorylated Ser526 in an in vitro kinase assay (Fig. 5K), suggesting that Aurora A is involved not only in Plk1 activation via Thr210 phosphorylation upon mitotic entry but also in mitotic catastrophe via Ser526 phosphorylation in mitotic cells harboring DNA damage. Strikingly, the interaction of the 3D mutant with Sgo1 substantially decreased in an in vitro pull-down assay (Fig. 5L), strongly suggesting that DDR-mediated phosphorylation of the PBD by Chk1 disrupts the interaction with Sgo1 and its translocation to the centrosome.
A–C After transfection of plasmids and B[a]P treatment, the localization of myc-Plk1 and multipolar spindles was analyzed (n = 300). D, E Chemically treated HeLa cells were analyzed by immunoblotting. F Cells were treated with the indicated inhibitors and B[a]P and analyzed by immunoblotting. G, H WT or mutant His-Plk1 was incubated with active Chk1 and analyzed by immunoblotting. I Recombinant Chk1 was incubated with recombinant His-Plk1 and proteins pulled down with Ni-beads were analyzed by immunoblotting. J After treatment with 0.5 µM B[a]P for 48 h or 300 ng/ml nocodazole for 16 h, cells were treated with 1 µM RO3306 as a Cdk1 inhibitor for 5 h, 1 µM BI2536 as a Plk1 inhibitor for 1 h, 5 µM VX680 as an Aurora A inhibitor for 1 h, and 2 µM Hesperadin as an Aurora B inhibitor for 5 h. Cell lysates were analyzed by immunoblotting. K His-Plk1 was incubated with Aurora A, pulled down with Ni+-beads, and analyzed by immunoblotting. L Lysates from DNA-transfected HeLa cells were incubated with recombinant His-Sgo1 and proteins pulled down with Ni-beads were analyzed by immunoblotting. Images of uncropped blots and gels are provided as a Supplementary Material file. Scale bar, 5 µm. Error bars, SEM. *p < 0.01 (two-tailed t test).
Hornerin forms a complex with Plk1 and Chk1 to mediate Plk1 phosphorylation
To delineate the mechanism underlying DDR-induced DiM, we next sought to identify a mediator between Chk1 and Plk1 through purification of the Plk1 complex and found hornerin via mass spectrometry (Fig. S5A, B). Analysis of a deletion mutant of hornerin revealed that Chk1 and Plk1 bound to the N-terminus and C-terminus of hornerin, respectively, to form an integrative complex between the DDR and DiM (Fig. S5C, D, E, F). Consistent with this idea, the interaction between Chk1 and Plk1 was abolished by hornerin-depletion (Fig. 6A). Furthermore, the levels of Plk1 and Sgo1 at the centrosome were recovered by the depletion of hornerin in B[a]P treated cells (Fig. 6B, C), presumably because hornerin mediates the phosphorylation of Plk1 by Chk1. Indeed, the phosphorylation of Plk1 at Ser529 and Thr539, but not Ser526, after B[a]P treatment was dramatically diminished in hornerin-depleted cells (Fig. 6D), suggesting that Ser529 and Thr539 are phosphorylated by Chk1 in a DDR- and hornerin-dependent manner but that Ser526 is phosphorylated by Aurora A in a DDR- and hornerin-independent manner. Consistently, overexpression of WT or siRNA-resistant hornerin (Myc-Hornerin-R) restored the ability of B[a]P-treated cells to form multipolar spindles in hornerin-depleted cells, as both siHornerin-A and siHornerin-B target the noncoding sequence of the endogenous hornerin gene (Fig. S6A, B, C, D, E). However, depletion of hornerin did not dissociate the Cep192::Aurora A::Plk1 complex (Fig. 6E). Therefore, DNA damage response did not interfere with the translocation of Cep192-AurA-Plk1 complex to the centrosome and could not induce the centriole disengagement in hornerin-depleted cells (Fig. 6B). Because DNA damage response could not eliminate mitotic cells with DNA damages through DiM without hornerin, depletion of hornerin increased the number of metaphase cells harboring damaged DNA by incapacitating DDR-induced DiM (Fig. S7). Intriguingly, the Plk1 3D mutant strongly interacted with hornerin in an in vitro pull-down assay, an immunoprecipitation assay, and proximity ligation assays (in situ PLAs) (Fig. 6F, G, H), possibly explaining the impaired translocation of Plk1 to the centrosome through trapping phosphorylated Plk1 by hornerin in the cytoplasm under B[a]P treatment. As expected, hornerin did not localize at the centrosome but exhibited diffusive localization in the cytoplasm (Fig. 6I). Hornerin-mediated multipolarity is not a cell type- or tissue-specific mechanism because similar centrosome perturbation by DDAs was observed in osteosarcoma epithelial cells, such as U2OS cells (Fig. S8A). Although skin cancer cells exhibited resistance against B[a]P probably because of their capacity of DNA repair, we also confirmed hornerin-mediated multipolarity in melanomas, including SK-MEL-2 and SK-MEL-28 cells, with etoposide (Fig. S8B, C).
A–D siRNA-transfected HeLa cells were treated with B[a]P and analyzed by IP-immunoblotting (A and E), immunostaining (B and C; n = 300), and immunoblotting (D). E siRNA-transfected HeLa cells were treated with B[a]P and analyzed by IP-immunoblotting. F Twenty-four hours after transfection of WT or mutant Plk1, the cell lysates were incubated with recombinant His-Plk1 and proteins pulled down with Ni-beads were analyzed by immunoblotting. G HeLa cells transfected with the indicated plasmid were subjected to IP-immunoblotting. H Hornerin-Plk1 3D proximity by in situ PLA in HeLa cells transfected with myc-Plk1 WT or mutants (n = 30). I HeLa cells were stained with hornerin (green), γ-tubulin (red), and DAPI (blue). Images of uncropped blots and gels are provided as a Supplementary Material file. AU, arbitrary units, Scale bars, 5 µm. Error bars, SEM. *p < 0.01 (two-tailed t test).
DDR inactivates Plk1 through Chk1-mediated phosphorylation of the PBD
Whereas the level of WT Plk1 was increased at the centrosome by neutralizing the sequestration of phosphorylated Plk1 through hornerin depletion, the level of the 3D mutant in the centrosome was not recovered in hornerin-depleted cells (Fig. 7A). Given that Cep192 recruits Plk1 and Aurora A to the centrosome [46] and that the kinase activity of Plk1 is pivotal for Cep192 targeting to the centrosome [47], we reasoned that the phosphorylation of the PBD by Chk1 might affect Plk1 activation. In agreement with our hypothesis, the levels of Cep192 and Aurora A in the centrosome were decreased in B[a]P treated cells (Fig. 7B, C). Furthermore, because the 3D mutant, similar to the WT and 3 A mutant, interacted with Cep192 (Fig. 6G), the level of Cep192 was decreased in the centrosome via sequestration with hornerin due to the dominant negative effect of the 3D mutant (Fig. 7D). In contrast, overexpression of the 3D mutant did not decrease the level of Sgo1, as endogenous Plk1 targets Sgo1 to the centrosome and, therefore, did not cause multipolar spindles (Figs. 7E, 5C). As both PBD docking-induced conformational changes and phosphorylation of Thr210 in the T-loop activate Plk1 [48], we examined the centrosome localization of T210A and 4D (T210D, S526D, S529D, and T539D) mutants to investigate the reason for the impaired targeting of the Plk1 3D mutant even in the absence of hornerin. Interestingly, both the T210A and 4D mutants exhibited half centrosome targeting and rescue capacity similar to that of WT (Fig. 7F, G). Furthermore, Thr210 in the 3D mutant was not phosphorylated (Fig. 7H), indicating that the phosphorylation of the PBD by the DDR completely disrupts the activation of Plk1 through conformational change and disturbing Thr210 phosphorylation. Indeed, centrosome separation in G2 was perturbed in DDA-treated cells (Fig. S9), which is reminiscent of the depletion of Plk1 protein or the inhibition of Plk1 activity. The delay in mitotic progression caused by perturbation of centrosome separation presumably induces DiM in cells harboring DNA damage and bipolar spindles (Fig. 1B). Because Ser137 and Trp414 are reportedly essential for Thr210 phosphorylation and centrosome targeting of Plk1 [49,50,51], we examined the interplay among these residues. While S137A or S137D mutants localized to the centrosome with perturbed centrosome integrity, the addition of a S137A mutation into the 4D mutant disrupted localization to the centrosome (Fig. S10A), suggesting that the phosphorylation of Ser137 is essential for the centrosome localization of Plk1 and the centrosome integrity. Next, we investigated whether the addition of S137D mutation into the 3D mutant rescues the centrosome localization of 3D mutant by inducing Thr210 phosphorylation. Unexpectedly, the S137D mutation in 3D mutant did not recover the Thr210 phosphorylation suppressed by 3D mutation and the centrosome localization of 3D mutant (Fig. S10A, B). These data indicate that phosphorylation of Ser137 is also required for the centrosomal function of Plk1 and that triple phosphorylation of PBD invalidates the function of phosphorylation at Ser137 for centrosome targeting of Plk1. Consistent with a previous report, the addition of the W414F mutation into the 4D mutant disrupted centrosome targeting of Plk1, suggesting that Trp414 is essential for centrosome targeting of Plk1 (Fig. S10A). Together, our results provide strong evidence that DDR-hornerin-mediated Plk1 phosphorylation at the PBD disturbs Plk1 activation and concurrent translocation of the Cep192::Aurora A::Plk1 complex and Sgo1 to the centrosome to induce spindle multipolarity and concomitant mitotic catastrophe.
A Twenty-four hours after siRNA-transfection, HeLa cells were transfected with the indicated plasmids and analyzed by immunostaining (n = 300). B, C HeLa cells were treated with B[a]P and analyzed by immunostaining (n = 300). D–G HeLa cells transfected with the indicated plasmid were analyzed by immunostaining (n = 300). H Twenty-eight hours after transfection of WT or mutant Plk1, the cells were treated with B[a]P for two hours and then analyzed by immunoblotting. Images of uncropped blots are provided as a Supplementary Material file. KD, kinase domain. AU, arbitrary units, Scale bar, 5 µm. Error bars, SEM. *p < 0.01 (two-tailed t test).
Hornerin governs DDR-induced DiM
We have described a molecular pathway leading to DiM via DDR. Our data also show a dissection between DiM with multipolar spindles and binucleation in mitotic catastrophe. Interestingly, the size of the colonies in the clonogenic assay was decreased by depletion of hornerin (Fig. 8A, B). Because individual phosphorylation at Ser526, Ser529 or Thr539 in PBD of Plk1 was detected in G2 or mitosis under unperturbed conditions (Fig. 5D, E), we propose that single or double phosphorylation in PBD is required for cell cycle progression. As DDR typically induces different types of cell death, including apoptosis, necroptosis, and mitotic catastrophe, in compliance with the type of damage and cell cycle stage [52, 53], we speculated that the level of hornerin-mediated DiM might not exceed the level of cell death in interphase. To directly test this hypothesis, we evaluated the proportion of cells that underwent hornerin-mediated DiM among all cells that underwent DDA-induced cell death. Strikingly, however, depletion of hornerin drastically increased the cell survival rate by over 50% compared to that of control cells treated with etoposide in long-term clonogenic assays (Fig. 8A, C). Consistent with this, the mRNA expression level of hornerin showed the strongest correlation with the survival rate of patients with cervical, pancreatic, or renal cancer (Fig. 8D and Fig. S11). Because depletion of hornerin specifically abrogated PBD phosphorylation in Plk1 but not active phosphorylation of Chk1 (Figs. 6D, 8E), we conclude that hornerin-mediated DiM is one of the major-types of DDR-induced cell death for eliminating damaged cells throughout the cell cycle.
A Colony formation assay (14 days) following hornerin knockdown. The image is representative of three independent experiments. B The formation of colonies by B[a]P (0.5 µM, replaced every three days) or etoposide (1 µM, replaced every three days) was quantified and plotted. C The size of colonies was quantified and plotted (n = 100 colonies from three independent experiments). D The survival curves of patients stratified according to the expression of hornerin mRNA in the TCGA database. Log-rank P value for Kaplan-Meier plot showing results from analysis of correlation between mRNA expression level and patient survival. E HeLa cells were treated with the indicated DDAs and analyzed by immunoblotting. AU, arbitrary units, Error bars, SEM. *p < 0.01 (two-tailed t test).
Discussion
Here, we identify the molecular mechanism by which hornerin, which is a component of the epidermal differentiation complex and contributes to envelope cornification and the epidermal antimicrobial barrier [54, 55], mediates the phosphorylation of the PBD in Plk1 by Chk1 to link DDR to DiM through premature centriole disengagement. We demonstrate that hornerin controls Plk1 and Sgo1 recruitment to centrosomes upon DNA damage by regulating Chk1-mediated phosphorylation of Plk1 (Fig. 6). Importantly, our mutational studies uncovered three critical residues in Plk1 required for the sequestration of Plk1 by hornerin, the intervention of Plk1 interaction with Sgo1, and the suppression of Plk1 activation by inhibiting T-loop phosphorylation and conformational change (Fig. 7F, G). Depletion of Plk1 and Sgo1 in centrosomes caused by Chk1-mediated Plk1 phosphorylation results in premature centriole disengagement in G2 and concurrent multipolar spindles in mitosis, leading to DiM.
Maintenance of genome stability, which is safeguarded by the precise orchestration of DNA replication, DNA repair, chromosome segregation, and cell cycle checkpoints, is the primary objective of cell cycle progression. Because DNA is replicated, compacted, and untangled through the cell cycle, chromatin, the substrate of DNA repair, undergoes structural transitions along with cell cycle progression. In this regard, the cell cycle restricts the type of DNA repair in distinct cell cycle phases. While nonhomologous end-joining (NHEJ) is the major repair pathway for DNA double strand breaks (DSBs) in G1, homologous recombination (HR) is restricted to the S and G2 phases because the ideal template of HR is the sister chromatid [56, 57]. Although mitotic cells permit minimal DNA repair for Holliday junction (HJ) dissolution and resolution [58], the major DNA repair pathways, including NHEJ and HR, are suppressed by Cdk1-mediated phosphorylation of the E3 ubiquitin ligases RNF8 and 53BP1 to prevent sister telomere fusions during mitosis [3]. However, mitotic cells sense all types of DSBs and initiate a limited DDR signaling cascade to mark their DNA damage for repair after mitotic exit [59]. Whereas NBS1 and Ku localize to mitotic DSBs and H2AX is phosphorylated in an ATM- and DNA-PK-dependent manner, the signaling cascade downstream of MDC1, including RNF8, RNF168, BRCA1, and 53BP1, fails to colocalize with γH2AX [60, 61]. In addition, Plk1 inhibits the DDR in mitosis by phosphorylating Chk2 and mediating the degradation of claspin, the adaptor protein of Chk1, through β-TrCP-SCF [62, 63]. Conversely, Chk1 phosphorylates Thr210 to activate Plk1 in unperturbed mitosis [43]. As an abrogated or compromised G2 checkpoint allows premature mitotic entry of defective cells harboring DNA damage or incompletely replicated DNA thereby leading to mitotic catastrophe [64,65,66], the crucial question arises as to how DDR induces DiM after entry into mitosis. DNA damage progressing from failed earlier checkpoints could provoke destructive cellular architecture during mitosis [26, 67, 68]. Our results, along with previous observations, indicate that premitotic DNA damage triggers DiM through ATR-Chk1-dependent premature centriole disengagement in G2 and concomitant multipolar spindle formation in mitosis (Fig. 3). Despite various inhibition of DNA repair and DDR, DNA damage induced in mitosis affects mitotic progression as severe DNA damage appears to induce a SAC-dependent but DDR-independent mitotic delay and concomitant mitotic catastrophe [69,70,71].
In addition to its primary function as a center of microtubule nucleation in both interphase and mitosis, centrosomes communicate with the DDR apparatus during cell cycle progression [18]. The initial activation of Cdk1, which occurs in the centrosome prior to the activation of nuclear Cdk1 and concomitant mitotic entry, is inhibited by Chk1- or Chk2-mediated phosphorylation of centrosomal CDC25 phosphatase upon DNA damage [72,73,74]. Furthermore, a number of DDR proteins, such as ATM, ATR, Chk1, Chk2, BRCA1, BRCA2, and poly(ADP-ribose) polymerases (PARPs), have been found at centrosomes during distinct phases of the cell cycle [18]. Thus, the centrosome is the center of cell fate determination throughout the cell cycle, as interdependent pathways of the DDR and cell cycle intersect at centrosomes. Our observations show that hornerin-mediated DiM, of which the mechanism is Plk1 phosphorylation by Chk1 and Aurora A and concurrent structural disintegration of the spindle, is the major pathway to prevent inheritance of damaged DNA in mitotic exit (Figs. 1B, 8C). In this context, the gain of centrosome clustering function endows cancer cells with the ability to escape DiM because supernumerary centrosomes are the most common feature in many tumor tissues, high-grade and recurrent tumors, and cancer cell lines [75,76,77]. Moreover, perturbed expression of hornerin possibly provides a starting point for the emergence of DDA-resistant cancer cells due to cancer-specific DDR defects, as depletion of hornerin substantially increased the survival rate of etoposide-treated cells (Fig. 8A, C). Thus, hornerin-mediated DiM, as the checkpoint in mitotic exit, is clinically relevant and could be exploited to improve therapeutic outcomes for cancer patients (Fig. 8D and Fig. S11).
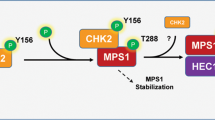
We established bifurcated pathways to induce mitotic catastrophe. The primary pathway is represented by hornerin which links the DDR to DiM by mediating the phosphorylation of Plk1 by Chk1 and the concurrent depletion of Sgo1 and the Cep192::Aurora A::Plk1 complex at the centrosome (Fig. 9). Cells with DNA damage but evading DiM are eradicated by Aurora B-mediated activation of the abscission checkpoint [7]. DDR-induced DiM, as the safeguard at the end of the cell cycle, ensures the faithful inheritance of genetic information to the next generation of cells.
Materials and methods
Plasmids
GFP-tagged Sgo1 expression vector, myc-tagged Separase expression vector, and pCer-C3-Plk1-K82R vector were purchased from Addgene (Watertown, MA). Point mutants of Plk1 (S526D, S529D, T539D, S526D/S529D, S526D/T539D, S529D/T539D, S526D/S529D/T539D, T210DS526D/S529D/S539D, T210A, S529A, T539A, or S526A/S529A/T539A) were generated using a Muta-directTM site-directed mutagenesis kit (iNtRON BIOTECHNOLOGY, Seongnam, Korea) according to the manufacturer’s protocol. To generate the His-Plk1 or His-Sgo1 vector, the PCR product of the human Plk1 or Sgo1 gene was cloned into the PET-28a vector (Novagen, Darmstadt, Germany).
Antibodies
Rabbit antibodies against phosphorylated Ser529 and phosphorylated Thr539 in Plk1 were generated using C-AIILHLSNG(p)SVQIN and QINFFQDH(p)TKLILC phosphor-peptides, respectively (AbClon, Seoul, Korea). Rabbit antibodies against phosphorylated Ser526 were generated using L(p)SNGSVQI-C phospho-peptide (GenScript, New Jersey, USA). For Western blotting, antibodies against the following were used (clone name, dilution, manufacturer and catalog number in parentheses): cyclin A (B-8, 1:1000, Santa Cruz Biotechnology Inc, sc-271682), cyclin B1 (H-20, 1:1000, Santa Cruz Biotechnology Inc., sc-594), p38 MAPK (N-20, 1:1000, Santa Cruz Biotechnology Inc., sc-728), Sgo1 (1:1000, Thermo Scientific Pierce Antibodies, PA5-30869), β-tubulin E7 monoclonal antibody (1:1000, Developmental Studies Hybridoma Bank USA, E7), separase (XJ11-1812, 1:500, Abcam, ab16170), PLK4 (6H5, 1:500, Millipore, MABC544), SAS-6 (91.390.21, 1:100, Santa Cruz Biotechnology Inc., sc-81431), STIL (1:500, Abcam, ab89314), Chk1 (2G1D5,1:2500, Cell Signaling Technology, #2360), c-Myc (9E10, 1:1000, Santa Cruz Biotechnology Inc., sc-40), HSP70 (3A3, 1:10000, Santa Cruz Biotechnology Inc., sc-32239), hornerin (1:500, NOVUS Biologicals, NBP1-80807), AhR (1:1000, Proteintech, 67785-1), purified Plk1 pT539 (1:500, AbClon), and purified Plk1 pS529 (1:500, AbClon). For immunostaining, antibodies against the following were used (clone name, dilution, manufacturer and catalog number in parentheses): Aurora B T232 (1:100, Rockland Immunochemicals, Inc., 600-401-677), Sgo1 (F-8, 1:100, Santa Cruz Biotechnology Inc., sc-393993), PLK1 (E-2, 1:100, Santa Cruz Biotechnology Inc., sc-55504), PLK4 (6H5, 1:500, Millipore, MABC544), SAS-6 (91.390.21, 1:100, Santa Cruz Biotechnology Inc., sc-81431), STIL (1:500, Abcam, ab89314), C-NAP1 (1:100, Proteintech, 14498-1-AP), VPS4B (UT292, 1:50, Sigma-Aldrich, ABS1656), ALIX (3A9, 1:100, Thermo Scientific Pierce Antibodies, MA1-83977), lamin B1 (EPR8985(B), 1:100, Abcam, ab194106), pericentrin (1:100, Proteintech, 22271-1-AP), β-tubulin E7 monoclonal antibody (1:100, Developmental Studies Hybridoma Bank USA, E7), γ-tubulin (GTU-88, 1:100, Sigma-Aldrich, T6557), c-Myc(9E10, 1:1000, Santa Cruz Biotechnology Inc., sc-40), centrin(1:100, Proteintech, 12794-1-AP), purified PLK1 pT539 (1:400, AbClon), purified PLK1 pS529 (1:200, AbClon), phospho-H2AX (1:200, Cell Signaling), phospho-MPM2 (1:1000, Millipore, 05-368), Alexa Fluor® 488 goat anti-rabbit IgG (1:100, Thermo Scientific Pierce Antibodies, A11034), Alexa Fluor® 594 goat anti-mouse IgG (1:100, Thermo Scientific Pierce Antibodies, A11032), and Alexa Fluor® 594 goat anti-rabbit IgG (1:100, Thermo Scientific Pierce Antibodies, A11037).
Cell culture and transfection
HeLa and RPE1 cells were obtained from the American Type Culture Collection (ATCC) and cultured in Dulbecco’s modified Eagle’s medium (DMEM, WelGENE Inc.) supplemented with 10% fetal bovine serum (FBS, Invitrogen), 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen). The cells were maintained at 37 °C in a humidified atmosphere containing 5% CO2. All cell lines tested negative for mycoplasma contamination by PCR. The Fingerprinting of all cell lines by ‘AmplFLSTR identifiler PCR Amplification kit’ was tested in Korean Cell Line Bank. siRNAs were transfected into HeLa cells using DharmaFect 1 (Dharmacon, Inc.). DNA transfection was performed using Lipofectamine 2000 (Invitrogen, USA) in accordance with the manufacturer’s instructions.
Cell treatments
To generate DNA damage, HeLa cells were treated with 0.5 µM B[a]P (Sigma) and 2.5 µM B[a]P-7,8-diol-9,10-epoxide (Sigma) for 48 h or 0.5 µM etoposide (Sigma) for 24 h. For the inhibition experiments, cells were incubated with the ATR inhibitor NU6027 (2 µM) (Sigma), the ATM inhibitor KU55933 (20 nM) (Sigma), (Enzo Life Sciences), the Chk1 inhibitor UCN-01 (300 nM) (Sigma), the Chk2 inhibitor (1 µM), and the DNA-PK inhibitor (0.5 µM) for 24 h. For synchronization in G2, cells were treated with the Cdk1 inhibitor RO3306 (1 µM) for 24 h. For inhibition of mitotic kinases, cells were treated with the Cdk1 inhibitor RO3306 (1 µM) for 5 h, the Plk1 inhibitor BI2536 (1 µM) for 1 h, the Aurora A inhibitor VX680 (5 µM) for 1 h, and the Aurora B inhibitor Hesperadin (2 µM) (Selleckem) for 5 h. Mitotic arrest was achieved by the treatment of 100 ng/ml nocodazole for 16 h.
siRNAs and primers
The control siRNA was 5’-CGTACGCGGAATACTTCGATT-3’. The following siRNA sequences were used: siHornerin-A: 5‘-GCAACAUGGUUCUACAUCAUU-3‘, siHornerin-B: 5‘-CUGUCUUUGGUCAACAUGAUU-3‘, siSeparase: 5′-CAAGGUUAGAUUUAAUCCUUU-3′, siPlk1: 5′-CCUUGAUGAAGAAGAUCAC-3′, siAhR: 5’-CGUUAGAUGUUCCUCUGUG-3’. Detailed information on the primers used in this study is provided in Supplementary Table S1.
Immunoprecipitation
Antibodies were conjugated to Affi-Prep Protein A beads (Bio-Rad Laboratories) at a concentration of 0.3 mg/ml. We lysed 1 × 107 cells in 1 ml NP-40 lysis buffer (50 mM HEPES, pH 7.4, 200 mM KCl, 0.3% NP-40, 10% glycerol, 1 mM EGTA, 1 mM MgCl2, 0.5 mM DTT, and 10 µg/ml each of leupeptin, pepstatin, and chymostatin). To preserve phosphorylation, lysis buffer was supplemented with 1 mM NaVO4 and 0.5 µM microcystin LR (Alexis Biochemicals). Corresponding lysates were centrifuged, incubated with protein A beads coupled with preimmune rabbit IgG at 4 °C for 1 h, and then incubated with protein A beads coupled with specific antibodies at 4 °C overnight. The beads were washed 5x with lysis buffer, boiled in Laemmli sample buffer for 3 min and resolved via SDS–polyacrylamide gel electrophoresis (PAGE).
Immunofluorescence and live cell imaging
HeLa cells on coverslips were fixed with methanol at −20 °C for 30 min. Alternatively, soluble proteins were extracted from the cells with BRB80-T buffer (80 mM PIPES, pH 6.8, 1 mM MgCl2, 5 mM EGTA and 0.5% Triton X-100) and then fixed with 4% paraformaldehyde for 15 min at room temperature. Fixed cells were permeabilized and blocked with PBS-BT (1 × PBS, 3% BSA, and 0.1% Triton X-100) for 30 min at room temperature. Coverslips were then incubated with primary and secondary antibodies diluted in PBS-BT. Images were acquired using ZEN2 software (Carl Zeiss, Germany) under a Zeiss Axiovert 200 M microscope with a 1.4 NA plan-Apo 100× oil immersion lens and an HRm CCD camera. Deconvoluted images were obtained and analyzed using AutoDeblur v9.1 and AutoVisualizer v9.1 (AutoQuant Imaging). The insets show single focal planes of the boxed regions. All images in a panel were acquired under a constant exposure time for all channels. To eliminate the size difference in each marked region, the average intensity was obtained from the selected area. Upon background subtraction, the average intensities of the desired channel were normalized against the average intensity of the corresponding markers or subtracted from the average background intensity value. For the fluorescence intensity plot, all intensities were normalized to those of the control and plotted as relative intensities.
For time-lapse microscopy, HeLa cells stably expressing GFP-H2B or GFP-centrin 2 were cultured in Leibovitz’s L-15 medium (Invitrogen) supplemented with 10% FBS (Invitrogen) and 2 mM L-glutamine (Invitrogen). The cells were placed in a sealed growth chamber that was heated to 37 °C and were observed using a Zeiss Axiovert 200 M microscope with a 20× lens. Images were acquired every three minutes for 5 h using AxioVision 4.8.2 (Carl Zeiss).
Bacterially expressed proteins
pET-28-vector encoded, His6-tagged Plk1 WT, S529A, T539A, 3 A, 3D, or Sgo1 were expressed individually in E. coli BL21 DE3 (Novagen). We lysed bacteria in lysis buffer (50 mM Na2HPO4, 100 mM NaCl, 10% glycerol, 10 mM imidazole), added Ni2 + -NTA-agarose (Qiagen), and washed beads with washing buffer (50 mM Na2HPO4, 100 mM NaCl, 10% glycerol, 20 mM imidazole). Corresponding proteins were purified from Ni2 + -NTA-agarose (Qiagen) according to standard procedures.
Pull-down assay
For the in vitro pull-down assay, 1 µg of each protein (His-Plk1 WT, 3 A, 3D, or His-Sgo1) was incubated with 20 µl of Ni2 + -NTA-agarose beads (Qiagen) for 1 h at 4 °C. After three washes with washing buffer (50 mM Na2HPO4, 100 mM NaCl, 10% glycerol, 20 mM imidazole), the protein-bead complex was incubated with the corresponding cell lysates (500 µg of total protein) for 1 h at 4 °C. After being washed with washing buffer three times, the beads were denatured by the addition of Laemmli sample buffer and boiling for 5 min at 95 °C. Samples were analyzed by immunoblotting.
In vitro kinase assay
For the Chk1 kinase assay, 200 ng of human recombinant Chk1 kinase (SignalChem) and Aurora A (purified from asynchronous sf9 cells) were incubated with 1 μg of His-Plk1 WT, T210A, S526A, S529A, or T539A mutant and 10 μM ATP in 50 μl of kinase buffer (25 mM Tris-HCl (pH 7.5), 2 mM DTT, 10 mM MgCl2, 5 mM β-glycerophosphate, and 0.1 mM Na3VO4) for 30 min at 37 °C. Incorporation of phosphate into His-Plk1 was visualized by SDS-PAGE and immunoblotting with specific phospho-antibodies.
Tandem purification and proteomics
HeLa S3 cells expressing GFP-S-Plk1 were generated as described previously [78]. The cells were lysed in 1 ml of NP-40 lysis buffer (50 mM Hepes, pH 7.4, 200 mM KCl, 0.3% NP-40, 10% glycerol, 1 mM EGTA, 1 mM MgCl2, 0.5 mM DTT, 1 mM NaVO4, 0.5 μM microcystin LR, 5 mM C4H7NaO2, and 10 μg/ml each of leupeptin, pepstatin, and chymostatin). Lysates were centrifuged and incubated with protein A beads coupled with anti-GFP antibodies at 4 °C overnight. Antibody beads were recovered by centrifugation, washed five times with lysis buffer in the presence of 500 mM KCl, washed twice with lysis buffer W/O protease inhibitors, and treated with TEV protease overnight to release S-Plk1 from protein A beads and GFP antibodies. Protease inhibitors were added to the supernatant and incubated with S-protein agarose beads (Sigma) for 3 h at 4 °C. The beads were washed 5x with lysis buffer in the presence of 0.5 mM DTT and incubated with urea elution buffer (50 mM Tris pH 8.5, 8 M urea) for 30 min at room temperature. Purified protein complexes were analyzed by mass spectrometry as described previously [78].
In situ PLA assay
Cells transfected with plasmid were blocked with Duolink blocking solution (Signa Aldrich) for 60 min at 37 °C. Afterward, the cells were incubated with Myc primary antibody (1:100), Hornerin primary antibody (1:100) for 30 min at 37 °C. The cells were washed in wash buffer A and incubated with PLUS and MINUS PLA probes (1:5) for 60 min at 37 °C. The cells were washed in wash buffer A and then incubated with the ligase solution for 30 min at 37 °C. The cells were again washed in wash buffer A and incubated with the polymerase solution for 100 min at 37 °C. Finally the cells were washed in wash buffer B for 20 min and then 0.01x buffer B for 4 min. After DAPI was added, the cells were mounted and imaged.
Centrosome preparation
Growing cells were incubated with culture media containing 10 mM nocodazole and 10 mM cytochalasin D for 1 h at 37 °C. Cells were harvested and lysed in a solution (1 mM HEPES pH 7.2, 0.5% NP-40, 0.5 mM MgCl2, 0.1% β-mercaptoethanol, 1 mM PMSF and protease inhibitors (aprotinin, leupeptin, pepstatin at 1 μg/mL)). Swollen nuclei and chromatin aggregates were removed by centrifugation at 3500 rpm for 10 min, and the supernatant was filtered through a nylon mesh. After adding 10 mM HEPES and 1 μg/mL DNase I, lysate was loaded on the 50% sucrose solution and centrosomes were sedimented on a sucrose cushion by centrifugation at 7500 rpm for 30 min. After removing 1/3 of the initial volume of the supernatant from the top, resuspended supernatant was loaded onto a sucrose gradient consisting of 3 ml of 70%, 3 ml of 50%, and 3 ml of 40% sucrose solution and centrifuged at 25000 rpm for 1 h. Fourteen fractions with 500 μl per fraction were collected from the bottom.
Colony formation assay
The cells were seeded into dishes at a density of 500 cells per dish and incubated for 2 weeks. The cell culture medium was refreshed every 3 days. After 2 weeks, the cells were stained with crystal violet (0.5 mg/ml, Santa Cruz Biotechnology Inc.) for 30 min on a shaker after washing with PBS. After 30 min, the cells were washed with PBS several times. The colonies were photographed and cell colonies with >50 cells or more were counted.
TCGA RNA-seq data
All TCGA RNA-seq data were acquired from the Human Protein Atlas website (https://www.proteinatlas.org/). Based on the FPKM (number fragments per kilobase of exon per million reads) value of each gene, patients were classified into two expression groups and the correlation between expression level and patient survival was examined. The survival outcomes of the two groups were compared by Kaplan–Meier survival analysis and log-rank tests. All statistical analyses were performed using IBM SPSS Statistics version 20.0 (IBM Inc., Armonk, NY, USA). All tests were two-sided, and a p-value of 0.05 was used as the cutoff value to determine statistical significance.
Statistical analysis
Statistical analyses were performed with GraphPad Prism software using two-way ANOVA with Bonferroni’s multiple comparisons test or a two-tailed t test as indicated in the figure legends. The error bars represent the standard errors of the mean (SEMs) of 3 independent experiments. A p-value of <0.01 (two-tailed) was considered statistically significant. For phenotype analysis in mitotic cell, we randomly analyzed 30 cells from three independent experiments. Because the percentage of mitotic cells in culture dish is about 1~2%, 10 cells are sufficient for each experiment. For quantitative analysis of immunostaining, the investigators were blinded to sample allocation.
Data availability
All relevant data are readily available from the authors.
References
Choi W, Lee ES. Therapeutic Targeting of DNA Damage Response in Cancer. Int J Mol Sci. 2022;23:1701.
Matthews HK, Bertoli C, de Bruin RAM. Cell cycle control in cancer. Nat Rev Mol Cell Biol. 2022;23:74–88.
Orthwein A, Fradet-Turcotte A, Noordermeer SM, Canny MD, Brun CM, Strecker J, et al. Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science. 2014;344:189–93.
Vitale I, Galluzzi L, Castedo M, Kroemer G. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat Rev Mol Cell Biol. 2011;12:385–92.
Gascoigne KE, Taylor SS. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer cell. 2008;14:111–22.
Lebrec V, Poteau M, Morretton JP, Gavet O. Chk1 dynamics in G2 phase upon replication stress predict daughter cell outcome. Dev Cell. 2022;57:638.
Huang XX, Tran T, Zhang LN, Hatcher R, Zhang PM. DNA damage-induced mitotic catastrophe is mediated by the Chk1-dependent mitotic exit DNA damage checkpoint. P Natl Acad Sci USA. 2005;102:1065–70.
Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Bio. 2007;8:451–63.
Pihan GA. Centrosome dysfunction contributes to chromosome instability, chromoanagenesis, and genome reprograming in cancer. Front Oncol. 2013;3:277.
Arquint C, Nigg EA. The PLK4-STIL-SAS-6 module at the core of centriole duplication. Biochem Soc T. 2016;44:1253–63.
Tsou MFB, Wang WJ, George KA, Uryu K, Stearns T, Jallepalli PV. Polo Kinase and Separase Regulate the Mitotic Licensing of Centriole Duplication in Human Cells. Dev Cell. 2009;17:344–54.
Hamasaki M, Matsumura S, Satou A, Takahashi C, Oda Y, Higashiura C, et al. Pregnenolone Functions in Centriole Cohesion during Mitosis. Chem Biol. 2014;21:1707–21.
Schockel L, Mockel M, Mayer B, Boos D, Stemmann O. Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat Cell Biol. 2011;13:966–U211.
Lukow DA, Sheltzer JM. Chromosomal instability and aneuploidy as causes of cancer drug resistance. Trends Cancer. 2022;8:43–53.
Kalkan BM, Ozcan SC, Quintyne NJ, Reed SL, Acilan C. Keep Calm and Carry on with Extra Centrosomes. Cancers (Basel). 2022;14:442.
Milunovic-Jevtic A, Mooney P, Sulerud T, Bisht J, Gatlin JC. Centrosomal clustering contributes to chromosomal instability and cancer. Curr Opin Biotechnol. 2016;40:113–8.
Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–9.
Mullee LI, Morrison CG. Centrosomes in the DNA damage response-the hub outside the centre. Chromosome Res. 2016;24:35–51.
Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–93.
Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr Biol. 2012;22:R966–980.
Petsalaki E, Zachos G. DNA damage response proteins regulating mitotic cell division: double agents preserving genome stability. FEBS J. 2020;287:1700–21.
Yang CY, Hao JW, Kong DJ, Cui XL, Zhang W, Wang HB, et al. ATM-mediated Mad1 Serine 214 phosphorylation regulates Mad1 dimerization and the spindle assembly checkpoint. Carcinogenesis. 2014;35:2007–13.
Kabeche L, Nguyen HD, Buisson R, Zou L. A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation. Science. 2018;359:108–14.
Kim EM, Burke DJ. DNA damage activates the SAC in an ATM/ATR-dependent manner, independently of the kinetochore. Plos Genet. 2008;4:e1000015.
Thompson R, Gatenby R, Sidi S. How Cells Handle DNA Breaks during Mitosis: Detection, Signaling, Repair, and Fate Choice. Cells-Basel. 2019;8:1049.
Stellas D, Souliotis VL, Bekyrou M, Smirlis D, Kirsch-Volders M, Degrassi F, et al. Benzo[a]pyrene-induced cell cycle arrest in HepG2 cells is associated with delayed induction of mitotic instability. Mutat Res-Fund Mol M. 2014;769:59–68.
Bourke E, Dodson H, Merdes A, Cuffe L, Zachos G, Walker M, et al. DNA damage induces Chk1-dependent centrosome amplification. Embo Rep. 2007;8:603–9.
Shiizaki K, Kawanishi M, Yagi T. Modulation of benzo[a]pyrene-DNA adduct formation by CYP1 inducer and inhibitor. Genes Environ: Off J Jpn Environ Mutagen Soc. 2017;39:14.
Maiato H, Logarinho E. Mitotic spindle multipolarity without centrosome amplification. Nat Cell Biol. 2014;16:386–U323.
Loffler H, Fechter A, Liu FY, Poppelreuther S, Kramer A. DNA damage-induced centrosome amplification occurs via excessive formation of centriolar satellites. Oncogene. 2013;32:2963–72.
Douthwright S, Sluder G. Link Between DNA Damage and Centriole Disengagement/Reduplication in Untransformed Human Cells. J Cell Physiol. 2014;229:1427–36.
Wilhelm T, Olziersky AM, Harry D, De Sousa F, Vassal H, Eskat A, et al. Mild replication stress causes chromosome mis-segregation via premature centriole disengagement. Nat Commun. 2019;10:3585.
Inanc B, Dodson H, Morrison CG. A centrosome-autonomous signal that involves centriole disengagement permits centrosome duplication in G2 phase after DNA damage. Mol Biol Cell. 2010;21:3866–77.
Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42.
Tang JB, Erikson RL, Liu XQ. Checkpoint kinase 1 (Chk1) is required for mitotic progression through negative regulation of polo-like kinase 1 (Plk1). P Natl Acad Sci USA. 2006;103:11964–9.
Waizenegger IC, Hauf S, Meinke A, Peters JM. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410.
Sumara I, Vorlaufer E, Stukenberg PT, Kelm O, Redemann N, Nigg EA, et al. The dissociation of cohesin from chromosomes in prophase is regulated by polo-like kinase. Mol Cell. 2002;9:515–25.
Wang X, Yang Y, Duan Q, Jiang N, Huang Y, Darzynkiewicz Z, et al. sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by PIk1. Dev Cell. 2008;14:331–41.
Mohr L, Buheitel J, Schockel L, Karalus D, Mayer B, Stemmann O. An Alternatively Spliced Bifunctional Localization Signal Reprograms Human Shugoshin 1 to Protect Centrosomal Instead of Centromeric Cohesin. Cell Rep. 2015;12:2156–68.
Seong YS, Kamijo K, Lee JS, Fernandez E, Kuriyama R, Miki T, et al. A spindle checkpoint arrest and a cytokinesis failure by the dominant-negative polo-box domain of Plk1 in U-2 OS cells. J Biol Chem. 2002;277:32282–93.
Elia AEH, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299:1228–31.
Kishi K, van Vugt MATM, Okamoto K, Hayashi Y, Yaffe MB. Functional Dynamics of Polo-Like Kinase 1 at the Centrosome. Mol Cell Biol. 2009;29:3134–50.
Adam K, Cartel M, Lambert M, David L, Yuan L, Besson A, et al. A PIM-CHK1 signaling pathway regulates PLK1 phosphorylation and function during mitosis. J Cell Sci. 2018;131:jcs213116.
Smits VA, Klompmaker R, Arnaud L, Rijksen G, Nigg EA, Medema RH. Polo-like kinase-1 is a target of the DNA damage checkpoint. Nat Cell Biol. 2000;2:672–6.
Seki A, Coppinger JA, Jang C-Y, Yates JR, Fang G. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Sci (N. Y, NY). 2008;320:1655–8.
Joukov V, Walter JC, De Nicolo A. The Cep192-Organized Aurora A-Plk1 Cascade Is Essential for Centrosome Cycle and Bipolar Spindle Assembly. Mol Cell. 2014;55:578–91.
Watanabe S, Meitinger F, Shiau AK, Oegema K, Desai A. Centriole-independent mitotic spindle assembly relies on the PCNT-CDK5RAP2 pericentriolar matrix. J Cell Biol. 2020;219:e202006010.
Xu J, Shen C, Wang T, Quan J. Structural basis for the inhibition of Polo-like kinase 1. Nat Struct Mol Biol. 2013;20:1047–53.
Tsvetkov L, Stern DF. Phosphorylation of Plk1 at S137 and T210 is inhibited in response to DNA damage. Cell Cycle. 2005;4:166–71.
Peng B, Shi RF, Bian J, Li YW, Wang PP, Wang HL, et al. PARP1 and CHK1 coordinate PLK1 enzymatic activity during the DNA damage response to promote homologous recombination-mediated repair. Nucl Acids Res. 2021;49:7554–70.
Garcia-Alvarez B, de Carcer G, Ibanez S, Bragado-Nilsson E, Montoya G. Molecular and structural basis of polo-like kinase 1 substrate recognition: Implications in centrosomal localization. P Natl Acad Sci USA. 2007;104:3107–12.
Wang JYJ. Cell Death Response to DNA Damage. Yale J Biol Med. 2019;92:771–9.
Matt S, Hofmann TG. The DNA damage-induced cell death response: a roadmap to kill cancer cells. Cell Mol Life Sci. 2016;73:2829–50.
Wu ZH, Meyer-Hoffert U, Reithmayer K, Paus R, Hansmann B, He YH, et al. Highly Complex Peptide Aggregates of the S100 Fused-Type Protein Hornerin Are Present in Human Skin. J Invest Dermatol. 2009;129:1446–58.
Wu ZH, Meyer-Hoffert U, Bartels J, He YH, Harder J, Schroeder JM. Antimicrobially active hornerin peptides as protective factors of healthy skin. J Invest Dermatol. 2007;127:S2–S2.
Ackerson SM, Romney C, Schuck PL, Stewart JA. To Join or Not to Join: Decision Points Along the Pathway to Double-Strand Break Repair vs. Chromosome End Protection. Front Cell Dev Biol. 2021;9:708763.
Arnoult N, Correia A, Ma J, Merlo A, Garcia-Gomez S, Maric M, et al. Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN. Nature. 2017;549:548–52.
Wyatt HD, Sarbajna S, Matos J, West SC. Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells. Mol Cell. 2013;52:234–47.
Heijink AM, Krajewska M, van Vugt MA. The DNA damage response during mitosis. Mutat Res. 2013;750:45–55.
Blackford AN, Stucki M. How Cells Respond to DNA Breaks in Mitosis. Trends Biochem Sci. 2020;45:321–31.
van Vugt MA, Gardino AK, Linding R, Ostheimer GJ, Reinhardt HC, Ong SE, et al. A mitotic phosphorylation feedback network connects Cdk1, Plk1, 53BP1, and Chk2 to inactivate the G(2)/M DNA damage checkpoint. PLoS Biol. 2010;8:e1000287.
Mamely I, van Vugt MATM, Smits VAJ, Semple JI, Lemmens B, Perrakis A, et al. Polo-like kinase-1 controls proteasome-dependent degradation of claspin during checkpoint recovery. Curr Biol. 2006;16:1950–5.
Mailand N, Bekker-Jensen S, Bartek J, Lukas J. Destruction of Claspin by SCFbetaTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol Cell. 2006;23:307–18.
Mackey MA, Zhang XF, Hunt CR, Sullivan SJ, Blum J, Laszlo A, et al. Uncoupling of M-phase kinase activation from the completion of S-phase by heat shock. Cancer Res. 1996;56:1770–4.
Chan TA, Hermeking H, Lengauer C, Kinzler KW, Vogelstein B. 14-3-3Sigma is required to prevent mitotic catastrophe after DNA damage. Nature. 1999;401:616–20.
Vakifahmetoglu H, Olsson M, Tamm C, Heidari N, Orrenius S, Zhivotovsky B. DNA damage induces two distinct modes of cell death in ovarian carcinomas. Cell Death Differ. 2008;15:555–66.
Hut HMJ, Lemstra W, Blaauw EH, van Cappellen GWA, Kampinga HH, Sibon OCM. Centrosomes split in the presence of impaired DNA integrity during mitosis. Mol Biol Cell. 2003;14:1993–2004.
Ingemarsdotter C, Keller D, Beard P. The DNA damage response to non-replicating adeno-associated virus: Centriole overduplication and mitotic catastrophe independent of the spindle checkpoint. Virology. 2010;400:271–86.
Nitta M, Kobayashi O, Honda S, Hirota T, Kuninaka S, Marumoto T, et al. Spindle checkpoint function is required for mitotic catastrophe induced by DNA-damaging agents. Oncogene. 2004;23:6548–58.
Dotiwala F, Harrison JC, Jain S, Sugawara N, Haber JE. Mad2 prolongs DNA damage checkpoint arrest caused by a double-strand break via a centromere-dependent mechanism. Curr Biol. 2010;20:328–32.
Mikhailov A, Cole RW, Rieder CL. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr Biol. 2002;12:1797–806.
Furnari B, Rhind N, Russell P. Cdc25 mitotic inducer targeted by chk1 DNA damage checkpoint kinase. Science. 1997;277:1495–7.
Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, et al. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–501.
Uto K, Inoue D, Shimuta K, Nakajo N, Sagata N. Chk1, but not Chk2, inhibits Cdc25 phosphatases by a novel common mechanism. EMBO J. 2004;23:3386–96.
Ryniawec JM, Rogers GC. Centrosome instability: when good centrosomes go bad. Cell Mol Life Sci. 2021;78:6775–95.
Sabat-Pospiech D, Fabian-Kolpanowicz K, Prior IA, Coulson JM, Fielding AB. Targeting centrosome amplification, an Achilles’ heel of cancer. Biochem Soc Trans. 2019;47:1209–22.
Sato N, Mizumoto K, Nakamura M, Maehara N, Minamishima YA, Nishio S, et al. Correlation between centrosome abnormalities and chromosomal instability in human pancreatic cancer cells. Cancer Genet Cytogen. 2001;126:13–19.
Jang CY, Wong J, Coppinger JA, Seki A, Yates JR 3rd, Fang G. DDA3 recruits microtubule depolymerase Kif2a to spindle poles and controls spindle dynamics and mitotic chromosome movement. J Cell Biol. 2008;181:255–67.
Acknowledgements
We thank Prof. Kunsoo Rhee (Seoul National University, Korea) for the cell line expressing GFP-centrin 2.
Funding
This work is supported by grants from the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (NRF-2020R1A2C1013663 and NRF-2022R1A5A2021216).
Author information
Authors and Affiliations
Contributions
All authors contributed to project planning and data analysis of their respective experiments. CYJ and GUB coordinated the work. HS, EHK, and JH performed all of the biochemical and imaging experiments. DG and JWK performed mutant analysis and interpreted the signaling pathway data. All authors contributed to the writing of the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Song, H., Kim, E.H., Hong, J. et al. Hornerin mediates phosphorylation of the polo-box domain in Plk1 by Chk1 to induce death in mitosis. Cell Death Differ 30, 2151–2166 (2023). https://doi.org/10.1038/s41418-023-01208-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-023-01208-y