Abstract
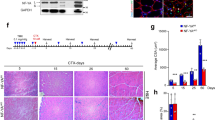
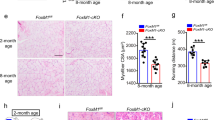
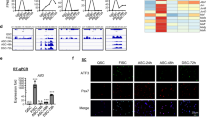
Skeletal muscle regeneration relies on muscle stem (satellite) cells. We previously demonstrated that satellite cells efficiently and accurately repair radiation-induced DNA double-strand breaks (DSBs) via the DNA-dependent kinase DNA-PKcs. We show here that DNA-PKcs affects myogenesis independently of its role in DSB repair. Consequently, this process does not require the accumulation of DSBs and it is also independent of caspase-induced DNA damage. We report that in myogenic cells DNA-PKcs is essential for the expression of the differentiation factor Myogenin in an Akt2-dependent manner. DNA-PKcs interacts with the p300-containing complex that activates Myogenin transcription. We show also that SCID mice that are deficient in DNA-PKcs, and are used for transplantation and muscle regeneration studies, display altered myofiber composition and delayed myogenesis upon injury. These defects are exacerbated after repeated injury/regeneration events resulting in reduced muscle size. We thus identify a novel, caspase-independent, regulation of myogenic differentiation, and define a differentiation phase that does not involve the DNA damage/repair process.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data analyzed during this study are included in this published article and the supplemental data files.
References
Vitale I, Manic G, De Maria R, Kroemer G, Galluzzi L. DNA damage in stem cells. Mol Cell. 2017;66:306–19. https://doi.org/10.1016/j.molcel.2017.04.006.
Zammit PS, Heslop L, Hudon V, Rosenblatt JD, Tajbakhsh S, Buckingham ME, et al. Kinetics of myoblast proliferation show that resident satellite cells are competent to fully regenerate skeletal muscle fibers. Exp Cell Res. 2002;281:39–49. https://doi.org/10.1006/excr.2002.5653.
Vahidi Ferdousi L, Rocheteau P, Chayot R, Montagne B, Chaker Z, Flamant P, et al. More efficient repair of DNA double-strand breaks in skeletal muscle stem cells compared to their committed progeny. Stem Cell Res. 2014;13:492–507. https://doi.org/10.1016/j.scr.2014.08.005.
Simonatto M, Marullo F, Chiacchiera F, Musaró A, Wang JY, Latella L, et al. DNA damage-activated ABL-MyoD signaling contributes to DNA repair in skeletal myoblasts. Cell Death Differ. 2013;20:1664–74. https://doi.org/10.1038/cdd.2013.118.
Larsen BD, Rampalli S, Burns LE, Brunette S, Dilworth FJ, Megeney LA. Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks. Proc Natl Acad Sci USA. 2010;107:4230–5. https://doi.org/10.1073/pnas.0913089107.
Chen X, Xu X, Chen Y, Cheung JC, Wang H, Jiang J, et al. Structure of an activated DNA-PK and its implications for NHEJ. Mol Cell. 2021;81:801–10.e803. https://doi.org/10.1016/j.molcel.2020.12.015.
Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell. 2017;66:801–17. https://doi.org/10.1016/j.molcel.2017.05.015.
Kotula E, Faigle W, Berthault N, Dingli F, Loew D, Sun JS, et al. DNA-PK target identification reveals novel links between DNA repair signaling and cytoskeletal regulation. PLoS ONE. 2013;8:e80313. https://doi.org/10.1371/journal.pone.0080313.
Matheny RW Jr., Geddis AV, Abdalla MN, Leandry LA, Ford M, McClung HL, et al. AKT2 is the predominant AKT isoform expressed in human skeletal muscle. Physiol Rep. 2018;6:e13652. https://doi.org/10.14814/phy2.13652.
Qian J, Wang Q, Dose M, Pruett N, Kieffer-Kwon KR, Resch W, et al. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 2014;159:1524–37. https://doi.org/10.1016/j.cell.2014.11.013.
Wang G, Zhu H, Situ C, Han L, Yu Y, Cheung TH, et al. p110alpha of PI3K is necessary and sufficient for quiescence exit in adult muscle satellite cells. EMBO J. 2018;37. https://doi.org/10.15252/embj.201798239.
Chen J, Wang Y, Hamed M, Lacroix N, Li Q. Molecular basis for the regulation of transcriptional coactivator p300 in myogenic differentiation. Sci Rep. 2015;5:13727. https://doi.org/10.1038/srep13727.
Knight JD, Kothary R. The myogenic kinome: protein kinases critical to mammalian skeletal myogenesis. Skelet Muscle. 2011;1:29. https://doi.org/10.1186/2044-5040-1-29.
Yoshida N, Yoshida S, Koishi K, Masuda K, Nabeshima Y. Cell heterogeneity upon myogenic differentiation: down-regulation of MyoD and Myf-5 generates ‘reserve cells’. J Cell Sci. 1998;111:769–79.
Puri PL, Bhakta K, Wood LD, Costanzo A, Zhu J, Wang JY. A myogenic differentiation checkpoint activated by genotoxic stress. Nat Genet. 2002;32:585–93. https://doi.org/10.1038/ng1023.
Zammit PS, Golding JP, Nagata Y, Hudon V, Partridge TA, Beauchamp JR. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J Cell Biol. 2004;166:347–57. https://doi.org/10.1083/jcb.200312007.
Abraham RT. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair. 2004;3:883–7. https://doi.org/10.1016/j.dnarep.2004.04.002.
Hunter T. When is a lipid kinase not a lipid kinase? When it is a protein kinase. Cell. 1995;83:1–4.
Bozulic L, Hemmings BA. PIKKing on PKB: regulation of PKB activity by phosphorylation. Curr Opin Cell Biol. 2009;21:256–61. https://doi.org/10.1016/j.ceb.2009.02.002.
Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, et al. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–51.
Serra C, Palacios D, Mozzetta C, Forcales SV, Morantte I, Ripani M, et al. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol Cell. 2007;28:200–13. https://doi.org/10.1016/j.molcel.2007.08.021.
Faralli H, Dilworth FJ. Turning on myogenin in muscle: a paradigm for understanding mechanisms of tissue-specific gene expression. Comp Funct Genomics. 2012;2012:836374. https://doi.org/10.1155/2012/836374.
Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. https://doi.org/10.1016/j.molcel.2010.09.019.
Tubbs A, Nussenzweig A. Endogenous DNA damage as a source of genomic instability in cancer. Cell. 2017;168:644–56. https://doi.org/10.1016/j.cell.2017.01.002.
Connolly PF, Fearnhead HO. DNA-PK activity is associated with caspase-dependent myogenic differentiation. FEBS J. 2016;283:3626–36. https://doi.org/10.1111/febs.13832.
Fukada S, Morikawa D, Yamamoto Y, Yoshida T, Sumie N, Yamaguchi M, et al. Genetic background affects properties of satellite cells and mdx phenotypes. Am J Pathol. 2010;176:2414–24. https://doi.org/10.2353/ajpath.2010.090887.
Carraro U, Dalla Libera L, Catani C. Myosin light and heavy chains in muscle regenerating in absence of the nerve: transient appearance of the embryonic light chain. Exp Neurol. 1983;79:106–17.
Brzoska E, Ciemerych MA, Przewozniak M, Zimowska M. Regulation of muscle stem cells activation: the role of growth factors and extracellular matrix. Vitam Horm. 2011;87:239–76. https://doi.org/10.1016/b978-0-12-386015-6.00031-7.
Bencze M, Negroni E, Vallese D, Yacoub-Youssef H, Chaouch S, Wolff A, et al. Proinflammatory macrophages enhance the regenerative capacity of human myoblasts by modifying their kinetics of proliferation and differentiation. Mol Ther. 2012;20:2168–79. https://doi.org/10.1038/mt.2012.189.
Panci G, Chazaud B. Inflammation during post-injury skeletal muscle regeneration. Semin Cell Dev Biol. 2021;119:32–38. https://doi.org/10.1016/j.semcdb.2021.05.031.
Hardy D, Besnard A, Latil M, Jouvion G, Briand D, Thepenier C, et al. Comparative study of injury models for studying muscle regeneration in mice. PLoS ONE. 2016;11:e0147198. https://doi.org/10.1371/journal.pone.0147198.
Röckl KS, Hirshman MF, Brandauer J, Fujii N, Witters LA, Goodyear LJ. Skeletal muscle adaptation to exercise training: AMP-activated protein kinase mediates muscle fiber type shift. Diabetes. 2007;56:2062–9. https://doi.org/10.2337/db07-0255.
Egawa T, Ohno Y, Goto A, Yokoyama S, Hayashi T, Goto K. AMPK mediates muscle mass change but not the transition of myosin heavy chain isoforms during unloading and reloading of skeletal muscles in mice. Int J Mol Sci. 2018;19. https://doi.org/10.3390/ijms19102954.
Bloemberg D, Quadrilatero J. Rapid determination of myosin heavy chain expression in rat, mouse, and human skeletal muscle using multicolor immunofluorescence analysis. PLoS ONE. 2012;7:e35273. https://doi.org/10.1371/journal.pone.0035273.
Kaneko S, Feldman RI, Yu L, Wu Z, Gritsko T, Shelley SA, et al. Positive feedback regulation between Akt2 and MyoD during muscle differentiation. Cloning of Akt2 promoter. J Biol Chem. 2002;277:23230–5. https://doi.org/10.1074/jbc.M201733200.
Vandromme M, Rochat A, Meier R, Carnac G, Besser D, Hemmings BA, et al. Protein kinase B beta/Akt2 plays a specific role in muscle differentiation. J Biol Chem. 2001;276:8173–9. https://doi.org/10.1074/jbc.M005587200.
Wilson EM, Tureckova J, Rotwein P. Permissive roles of phosphatidyl inositol 3-kinase and Akt in skeletal myocyte maturation. Mol Biol Cell. 2004;15:497–505. https://doi.org/10.1091/mbc.E03-05-0351.
Hoxhaj G, Manning BD. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat Rev Cancer. 2020;20:74–88. https://doi.org/10.1038/s41568-019-0216-7.
Bozulic L, Surucu B, Hynx D, Hemmings BA. PKBalpha/Akt1 acts downstream of DNA-PK in the DNA double-strand break response and promotes survival. Mol Cell. 2008;30:203–13. https://doi.org/10.1016/j.molcel.2008.02.024.
Liu P, Gan W, Guo C, Xie A, Gao D, Guo J, et al. Akt-mediated phosphorylation of XLF impairs non-homologous end-joining DNA repair. Mol Cell. 2015;57:648–61. https://doi.org/10.1016/j.molcel.2015.01.005.
Yue X, Bai C, Xie D, Ma T, Zhou PK. DNA-PKcs: a multi-faceted player in DNA damage response. Front Genet. 2020;11:607428. https://doi.org/10.3389/fgene.2020.607428.
Damia G. Targeting DNA-PK in cancer. Mutat Res. 2020;821:111692. https://doi.org/10.1016/j.mrfmmm.2020.111692.
Bustin M, Catez F, Lim JH. The dynamics of histone H1 function in chromatin. Mol Cell. 2005;17:617–20. https://doi.org/10.1016/j.molcel.2005.02.019.
Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–802. https://doi.org/10.1126/science.1127196.
Bouquet F, Ousset M, Biard D, Fallone F, Dauvillier S, Frit P, et al. A DNA-dependent stress response involving DNA-PK occurs in hypoxic cells and contributes to cellular adaptation to hypoxia. J Cell Sci. 2011;124:1943–51. https://doi.org/10.1242/jcs.078030.
Wrann S, Kaufmann MR, Wirthner R, Stiehl DP, Wenger RH. HIF mediated and DNA damage independent histone H2AX phosphorylation in chronic hypoxia. Biol Chem. 2013;394:519–28. https://doi.org/10.1515/hsz-2012-0311.
Fernando P, Kelly JF, Balazsi K, Slack RS, Megeney LA. Caspase 3 activity is required for skeletal muscle differentiation. Proc Natl Acad Sci USA. 2002;99:11025–30. https://doi.org/10.1073/pnas.162172899.
Murray TV, McMahon JM, Howley BA, Stanley A, Ritter T, Mohr A, et al. A non-apoptotic role for caspase-9 in muscle differentiation. J Cell Sci. 2008;121:3786–93. https://doi.org/10.1242/jcs.024547.
Al-Khalaf MH, Blake LE, Larsen BD, Bell RA, Brunette S, Parks RJ, et al. Temporal activation of XRCC1-mediated DNA repair is essential for muscle differentiation. Cell Discov. 2016;2:15041. https://doi.org/10.1038/celldisc.2015.41.
Wong RH, Chang I, Hudak CS, Hyun S, Kwan HY, Sul HS. A role of DNA-PK for the metabolic gene regulation in response to insulin. Cell. 2009;136:1056–72. https://doi.org/10.1016/j.cell.2008.12.040.
Taccioli GE, Amatucci AG, Beamish HJ, Gell D, Xiang XH, Torres Arzayus MI, et al. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity. 1998;9:355–66. https://doi.org/10.1016/s1074-7613(00)80618-4.
Anne Esguerra Z, Watanabe G, Okitsu CY, Hsieh CL, Lieber MR. DNA-PKcs chemical inhibition versus genetic mutation: Impact on the junctional repair steps of V(D)J recombination. Mol Immunol. 2020;120:93–100. https://doi.org/10.1016/j.molimm.2020.01.018.
Murphy MM, Keefe AC, Lawson JA, Flygare SD, Yandell M, Kardon G. Transiently active Wnt/beta-catenin signaling is not required but must be silenced for stem cell function during muscle regeneration. Stem Cell Rep. 2014;3:475–88. https://doi.org/10.1016/j.stemcr.2014.06.019.
Bosma GC, Custer RP, Bosma MJ. A severe combined immunodeficiency mutation in the mouse. Nature. 1983;301:527–30.
Beamish HJ, Jessberger R, Riballo E, Priestley A, Blunt T, Kysela B, et al. The C-terminal conserved domain of DNA-PKcs, missing in the SCID mouse, is required for kinase activity. Nucleic Acids Res. 2000;28:1506–13.
Araki R, Fujimori A, Hamatani K, Mita K, Saito T, Mori M, et al. Nonsense mutation at Tyr-4046 in the DNA-dependent protein kinase catalytic subunit of severe combined immune deficiency mice. Proc Natl Acad Sci USA. 1997;94:2438–43.
Blunt T, Gell D, Fox M, Taccioli GE, Lehmann AR, Jackson SP, et al. Identification of a nonsense mutation in the carboxyl-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse. Proc Natl Acad Sci USA. 1996;93:10285–90.
Pisciotta A, Riccio M, Carnevale G, Lu A, De Biasi S, Gibellini L, et al. Stem cells isolated from human dental pulp and amniotic fluid improve skeletal muscle histopathology in mdx/SCID mice. Stem Cell Res Ther. 2015;6:156. https://doi.org/10.1186/s13287-015-0141-y.
Grabowska I, Mazur MA, Kowalski K, Helinska A, Moraczewski J, Streminska W, et al. Progression of inflammation during immunodeficient mouse skeletal muscle regeneration. J Muscle Res Cell Motil. 2015;36:395–404. https://doi.org/10.1007/s10974-015-9433-1.
Farini A, Meregalli M, Belicchi M, Battistelli M, Parolini D, D’Antona G, et al. T and B lymphocyte depletion has a marked effect on the fibrosis of dystrophic skeletal muscles in the scid/mdx mouse. J Pathol. 2007;213:229–38. https://doi.org/10.1002/path.2213.
Gayraud-Morel B, Chretien F, Jory A, Sambasivan R, Negroni E, Flamant P, et al. Myf5 haploinsufficiency reveals distinct cell fate potentials for adult skeletal muscle stem cells. J Cell Sci. 2012;125:1738–49. https://doi.org/10.1242/jcs.097006.
Gayraud-Morel B, Chretien F, Flamant P, Gomes D, Zammit PS, Tajbakhsh S. A role for the myogenic determination gene Myf5 in adult regenerative myogenesis. Dev Biol. 2007;312:13–28. https://doi.org/10.1016/j.ydbio.2007.08.059.
Gayraud-Morel B, Pala F, Sakai H, Tajbakhsh S. Isolation of muscle stem cells from mouse skeletal muscle. Methods Mol Biol. 2017;1556:23–39. https://doi.org/10.1007/978-1-4939-6771-1_2.
Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–7.
Pinset C, Montarras D, Chenevert J, Minty A, Barton P, Laurent C, et al. Control of myogenesis in the mouse myogenic C2 cell line by medium composition and by insulin: characterization of permissive and inducible C2 myoblasts. Differentiation. 1988;38:28–34.
Rothkamm K, Lobrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci USA. 2003;100:5057–62. https://doi.org/10.1073/pnas.0830918100.
Kwon M, Firestein BL. DNA transfection: calcium phosphate method. Methods Mol Biol. 2013;1018:107–10. https://doi.org/10.1007/978-1-62703-444-9_10.
Dentice M, Ambrosio R, Damiano V, Sibilio A, Luongo C, Guardiola O, et al. Intracellular inactivation of thyroid hormone is a survival mechanism for muscle stem cell proliferation and lineage progression. Cell Metab. 2014;20:1038–48. https://doi.org/10.1016/j.cmet.2014.10.009.
Baghdadi MB, Castel D, Machado L, Fukada SI, Birk DE, Relaix F, et al. Reciprocal signalling by Notch-Collagen V-CALCR retains muscle stem cells in their niche. Nature. 2018;557:714–8. https://doi.org/10.1038/s41586-018-0144-9.
Crochemore C, Fernandez-Molina C, Montagne B, Salles A, Ricchetti M. CSB promoter downregulation via histone H3 hypoacetylation is an early determinant of replicative senescence. Nat Commun. 2019;10:5576. https://doi.org/10.1038/s41467-019-13314-y.
Urbani L, Piccoli M, Franzin C, Pozzobon M, De Coppi P. Hypoxia increases mouse satellite cell clone proliferation maintaining both in vitro and in vivo heterogeneity and myogenic potential. PLoS ONE. 2012;7:e49860. https://doi.org/10.1371/journal.pone.0049860.
Tintignac LA, Sirri V, Leibovitch MP, Lecluse Y, Castedo M, Metivier D, et al. Mutant MyoD lacking Cdc2 phosphorylation sites delays M-phase entry. Mol Cell Biol. 2004;24:1809–21.
Londhe P, Davie JK. Sequential association of myogenic regulatory factors and E proteins at muscle-specific genes. Skelet Muscle. 2011;1:14. https://doi.org/10.1186/2044-5040-1-14.
Acknowledgements
We thank the lab of Shahragim Tajbakhsh for the material provided to perform in vivo muscle injuries and antibodies targeting of myogenic factors, Hiroshi Sakai for guidance to initate the in vivo injury experiments, and Shahragim Tajbakhsh for helpful discussion, Sebastien Mella for his advice on statistical analysis. We thank the Virus and Immunity Laboratory at Institut Pasteur (director Dr. Schwartz) for quantification of p24 viral particles, the components of Elisa Perdiguero-Gomez lab at Institut Pasteur, in particular Alina Sommer, for the technical help with the analysis of macrophage immunolabeling, and the center for Translational Science (CRT)- Cytometry and Biomarkers Unit and Photonic BioImaging Unit of Technology and Service (CBUTechS and PBI UTechS) at Institut Pasteur for technical support in this study.
Funding
This work was supported by AFM (research grant (16580), and thesis grant (18425)).
Author information
Authors and Affiliations
Contributions
HHS planned, analyzed, and performed all experiments, except the immunostaining of macrophages, fiber composition (slow and fast fibers), and some experiments with the caspase inhibitor, which have been performed by BM. HHS also contributed to writing the manuscript. MR supervised the study, analyzed the data, and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sutcu, H.H., Montagne, B. & Ricchetti, M. DNA-PKcs regulates myogenesis in an Akt-dependent manner independent of induced DNA damage. Cell Death Differ 30, 1900–1915 (2023). https://doi.org/10.1038/s41418-023-01177-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-023-01177-2
This article is cited by
-
Isolation, identification, and induced differentiation of satellite cells from skeletal muscle of adult tree shrews
In Vitro Cellular & Developmental Biology - Animal (2024)