Abstract
Metabolic switch is critical for cell fate determination through metabolic functions, epigenetic modifications, and gene expression. However, the mechanisms underlying these alterations and their functional roles remain unclear. Here, we show that Plin2-mediated moderate lipid hydrolysis is critical for pluripotency of embryonic stem cells (ESCs). Upon exit from pluripotency, lipid droplet (LD)-associated protein Plin2 is recognized by Hsc70 and degraded via chaperone-mediated autophagy to facilitate LD mobilization. Enhancing lipid hydrolysis by Plin2 knockout promotes pluripotency exit, which is recovered by ATGL inhibition. Mechanistically, excessive lipid hydrolysis induces a dramatic lipidomic remodeling characterized by decreased cardiolipin and phosphatidylethanolamine, which triggers defects in mitochondrial cristae and fatty acid oxidation, resulting in reduced acetyl-CoA and histone acetylation. Our results reveal how LD mobilization is regulated and its critical role in ESC pluripotency, and indicate the mechanism linking LD homeostasis to mitochondrial remodeling and epigenetic regulation, which might shed light on development and diseases.
Similar content being viewed by others
Introduction
Pluripotent stem cells (PSCs) hold great promise for regenerative medicine and developmental biology studies since they have the unique ability to indefinitely self-renew and to differentiate into almost any cell type of the adult organism. The developmental plasticity of PSCs is associated with open and dynamic chromatin structure and a distinct metabolic state characterized by high glycolytic activity and lipid unsaturation [1, 2]. A pronounced metabolic switch from glycolysis to oxidative phosphorylation occurs during the loss of pluripotency, while the acquisition of pluripotency is accompanied by an opposite metabolic transition [3, 4]. Emerging evidence shows that metabolic switch is critical for stem cell fate determination not only through direct metabolic functions, but also by regulating epigenetic modifications and gene expression [4,5,6,7,8,9,10,11]. Lipids have been suggested to act as a carbon source for generating acetyl-CoA for histone acetylation [12, 13]. However, whether and how lipids are involved in epigenetic modulation of pluripotency remain unclear.
The metabolic switch during cell fate transitions is closely linked to the remodeling of cellular organelles. As a central metabolic organelle, mitochondria undergo dramatic morphological changes during the acquisition and loss of pluripotency, and the transitions between different pluripotent states, i.e., the naive and primed states [14]. Mitochondria in PSCs exhibit an immature morphology with globular shape and poorly developed cristae, which turn into a mature form with elongated appearance and well-developed cristae during early differentiation [14]. Surprisingly, compared to mitochondria in naive PSCs, mitochondria in primed PSCs are elongated with relatively more developed cristae, despite showing significantly reduced mitochondrial respiration, suggesting a complex coordination pattern between mitochondrial structure and function [15, 16]. The remodeling of organelles is primarily controlled by the cooperation between membrane proteins and lipids. Although protein machinery controlling mitochondrial remodeling in PSCs has been extensively studied, the regulatory roles of lipids remain largely obscure.
Lipid droplets (LDs) are key organelles involved in lipid storage and utilization, which consist of a neutral lipid core surrounded by a phospholipid monolayer and numerous proteins such as the perilipins. LDs are increasingly recognized as dynamic and multifunctional organelles that play an active role in a variety of physiological and pathological processes, such as neurodegeneration, tumorigenesis, and embryonic development [17,18,19,20,21,22,23,24,25]. LDs are highly abundant in mammalian oocytes and embryos and change dynamically during embryonic development [26]. Recent findings indicate that LDs are crucial for early embryonic development [20, 21]. Importantly, LDs accumulate during the process of pluripotency acquisition and their abundance varies in different pluripotent states [16], suggesting potential roles for LD homeostasis in pluripotency acquisition and transitions. LD homeostasis is maintained by the balance between lipid synthesis and hydrolysis. While lipid synthesis has been suggested to play important roles in pluripotency acquisition [8, 27, 28], less is known about lipid hydrolysis. How lipid hydrolysis is regulated and its functional role in cell fate decisions of PSCs remain largely unexplored.
Here, we show Plin2 safeguards pluripotency of mESCs by suppressing LD mobilization, which is crucial for maintaining mitochondrial cristae structure and histone acetylation. Upon exit from pluripotency, LD mobilization is upregulated by chaperone-mediated autophagy (CMA)-mediated degradation of Plin2 to facilitate differentiation. Enhancing lipid hydrolysis by Plin2 knockout induces lipidomic remodeling associated with disorganized mitochondrial cristae, thus leading to impaired mitochondrial respiration and reduced acetyl-CoA. The decrease in acetyl-CoA reduces acetylation of histone H3 at lysine 27 (H3K27ac) and promotes the exit from pluripotency. Together, these results reveal an important role for LD mobilization in pluripotency regulation, and provide insights into mechanisms of organelle remodeling in cell fate decisions.
Results
Plin2-dependent lipid droplets delay exit from pluripotency
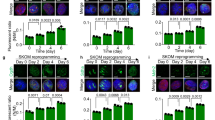
LDs are coated with one or more of five members of the perilipin family of proteins: Plin1-Plin5, which are the major structural proteins on LDs and play crucial roles in the formation and stabilization of LDs [29]. The perilipin family proteins vary by tissue and between different conditions [29, 30] (Fig. 1a). ESCs have been reported to possess LDs [16] (Supplementary Fig. 1a), but which Plin protein is associated with pluripotency remains unknown. We analyzed the expression of the five perilipin genes in mouse embryonic fibroblasts (MEFs) and embryonic stem cells (mESCs). Plin2 was highly expressed in mESCs, while Plin3 was mainly expressed in MEFs, suggesting Plin2 might play a role in pluripotency regulation (Fig. 1b). We therefore examined the expression of Plin2, Plin3, and the pluripotency marker, Oct4, during reprogramming of MEFs and spontaneous differentiation of mESCs. Plin2 expression was positively correlated with the expression of pluripotency gene Oct4, whereas Plin3 was negatively correlated with Oct4 (Fig. 1c). Similar results were obtained at the protein level by western blot of Plin2 and Oct4 (Fig. 1d). We further analyzed the location of Plin2 during reprogramming and differentiation and found circular structures of Plin2 that appeared upon reprogramming and disappeared during differentiation (Fig. 1e). In addition, knockdown of Plin2 suppressed the reprogramming of MEFs to a pluripotent state (Supplementary Fig. 1b). LDs are organelles that store neutral lipids, we then quantified neutral lipids, such as triglycerides (TGs), using the neural lipid dye BODIPY 493/503 by flow cytometry. The fluorescence intensity of BODIPY 493/503 increased markedly during reprogramming while decreased upon differentiation, indicating Plin2-positive LDs are associated with the accumulation of neutral lipid in PSCs (Fig. 1f).
a Schematic demonstrating the structure of lipid droplets and the main tissue distribution of perilipin family proteins: WAT white adipose tissue, BAT brown adipose tissue, MSC mesenchymal stem cell. b qRT-PCR analysis of expression of perilipin genes (Plin1, Plin2, Plin3, Plin4, Plin5) in MEF and mESC. Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. ***P < 0.001, **P < 0.005. c qRT-PCR analysis of expression of Plin2, Plin3 and Oct4 during reprogramming of MEFs and spontaneous differentiation of mESCs. Data are mean ± SD, n = 3 biological replicates. d Western blot analysis of Plin2 and Oct4 during reprogramming of MEFs and spontaneous differentiation of mESCs. The experiments were repeated independently three times with similar results. e Immunofluorescence staining of Plin2 (Red) during reprogramming of MEFs and spontaneous differentiation of mESCs. Scale bar, 25 μm. The experiments were repeated independently three times with similar results. f Representative histograms and MFI values of BODIPY 493/503 during reprogramming of MEFs and spontaneous differentiation of mESCs. Data are mean ± SD, n = 3 biological replicates. Two-tailed paired t-tests. ***P < 0.001, **P < 0.005, *P < 0.05. g Schematic depicting experimental design to reduce lipid droplets in mESCs and western blot analysis of Plin2 in WT and Plin2-/- mESCs (KO1 and KO2). The experiments were repeated independently twice with similar results. h Representative images of Oil Red O staining in WT and Plin2-/- mESCs (KO1 and KO2). Scale bar, 100 μm. The experiments were repeated independently three times with similar results. i Triglyceride (TG) content in WT and Plin2-/- mESCs (KO1 and KO2). Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. *P < 0.05. j Representative phase-contrast images and the expression of genes associated with naive pluripotency (Rex1, Tbx3, Klf4) and primed pluripotency (Fgf51, Otx2, Foxa2) of WT and Plin2-/- mESCs (KO1 and KO2) during naive-primed transition on day 5. Scale bar, 100 μm. Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. ***P < 0.001. k Representative phase-contrast and Oct4-GFP images and the expression of pluripotency markers (Oct4, Nanog, Rex1) of WT and Plin2-/- mESCs (KO1 and KO2) on day 3 of differentiation. Scale bar, 100 μm. Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. ***P < 0.001.
To gain insights into the possible roles of LDs in pluripotency regulation, we sought to reduce LD content in mESCs. We generated Plin2 knockout mESC clones (KO1 and KO2) by CRISPR/Cas9-mediated gene editing using two different sgRNAs (Fig. 1g and Supplementary Fig. 1c). Oil Red O staining of LDs showed that Plin2 knockout resulted in a remarkable reduction in the number of LDs in mESCs, which was accompanied by decreased TG content, suggesting Plin2 is essential for LD formation or stabilization (Fig. 1h, i). Plin2-/- mESCs showed mildly decreased proliferation (Supplementary Fig. 1d), and had typical undifferentiated morphology and expressed pluripotency genes (Fig. 1h and Supplementary Fig. 1e). Because LDs are differentially distributed in different pluripotent states [16], we sought to investigate whether LDs contribute to the transition between different states by inducing naive mESCs into primed mESCs. After 5 days of induction, wild-type (WT) mESCs turned into primed state and displayed a flattened morphology, while Plin2-/- mESCs showed spontaneous differentiation, indicated by the decrease of naive genes (Rex1, Tbx3, and Klf4) and of primed genes (Fgf5, Otx2, and Foxa2) (Fig. 1j). These results suggest that Plin2-dependent LDs are either required for the maintenance of primed pluripotency or can counteract differentiation.
We next examined the pluripotency maintaining capacity of Plin2-/- mESCs using an in vitro spontaneous embryoid body (EB) differentiation approach. Plin2-/- mESCs rapidly lost Oct4 expression and typical ESC morphology on day 3 of differentiation, while WT mESCs maintained a comparatively undifferentiated state (Fig. 1k). qRT-PCR analysis showed that the expression of pluripotency-associated genes was significantly decreased in Plin2-/- mESCs compared with WT mESCs upon exit from pluripotency (Fig. 1k) and the percentage of Oct4 positive cells, measured by flow cytometry, was reduced (Supplementary Fig. 1f). We also investigated the differentiation bias of Plin2-/- mESCs using qRT-PCR and found that markers of the three germ layers were increased in Plin2-/- mESCs compared with WT mESCs, suggesting Plin2 deficiency promotes spontaneous differentiation of mESCs into all three germ layers (Supplementary Fig. 1g). Western blot showed markers of ectoderm (beta-III tubulin), mesoderm (Brachyury), and endoderm (AFP) were all expressed in the late stage of differentiation (day 9), indicating Plin2 deficiency does not affect differentiation potential (Supplementary Fig. 1h). Together, these results demonstrate that loss of Plin2 lead mESCs prone to exit from pluripotency and indicate a role for LDs in pluripotency maintenance.
Plin2 safeguards pluripotency by suppressing lipid hydrolysis
Plin2 plays important roles in cytosolic lipolysis by acting as a structural barrier that protects LDs from lipases [31]. The observations that neural lipid decreases during differentiation and Plin2 deficiency accelerates exit from pluripotency promoted us to ask whether lipid hydrolysis contributes to the pluripotency exit of mESCs. We measured the relative lipid hydrolysis rate in undifferentiated mESCs and during the differentiation of mESCs by quantifying the remaining neural lipid and TG content after Triacsin C treatment to inhibit TG synthesis in a lipid-depleted medium [32] (Fig. 2a). The remaining neural lipid and TG content, quantified by BODIPY 493/503 staining and an enzymatic assay, respectively, decreased faster after Triacsin C treatment in mESCs on day 3 of differentiation compared with undifferentiated mESCs (Fig. 2a, b), which indicates lipid hydrolysis rate is increased upon exit from pluripotency. Notably, Plin2 deficiency resulted in a significant increase in lipid hydrolysis rate in mESCs (Fig. 2c), implying increased lipid hydrolysis may be responsible for the pluripotency exit of Plin2-/- mESCs. Thus, we tested whether inhibition of lipid hydrolysis could rescue Plin2 deficiency-induced differentiation using a selective inhibitor of adipose triglyceride lipase (ATGL), Atglistatin. Notably, ATGL inhibition by Atglistatin significantly decreased lipid hydrolysis in a dose-dependent manner (Supplementary Fig. 2a), and retarded the pluripotency exit of Plin2-/- mESCs, while had little effect on WT mESCs, evidenced by the percentage of Oct4-GFP positive cells, pluripotency gene expression, and relative undifferentiated morphology (Fig. 2d, e and Supplementary Fig. 2b). These data demonstrate that Plin2 deficiency accelerates exit from pluripotency by enhancing lipid hydrolysis.
a Representative histograms and MFI values of BODIPY 493/503 of undifferentiated mESCs (day 0) and differentiating mESCs (day 3) at the indicated time points during the hydrolysis assay as shown at the top left. Data are mean ± SD, n = 3 biological replicates. Two-tailed paired t-tests. ***P < 0.001. b TG content in undifferentiated mESCs (day 0) and differentiating mESCs (day 3) at the indicated time points during the hydrolysis assay. Remaining TG is expressed as a percentage of the TG content at 0 h. Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. ***P < 0.001, *P < 0.05. c TG content in WT and Plin2-/- mESCs (KO1 and KO2) at the indicated time points during the hydrolysis assay. Remaining TG is expressed as a percentage of the TG content at 0 h. Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. ***P < 0.001, **P < 0.005, *P < 0.05. d Flow cytometry analysis and quantification of Oct4-GFP+ cells in WT and Plin2-/- mESCs (KO1 and KO2) on day 3 of differentiation treated with DMSO or ATGLi (10 μM Atglistatin). Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. ***P < 0.001. e qRT-PCR analysis of expression of pluripotency markers (Oct4, Nanog, Rex1) in WT and Plin2-/- mESCs (KO1 and KO2) on day 3 of differentiation treated with DMSO or ATGLi (10 μM Atglistatin). Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. ***P < 0.001, **P < 0.005.
Plin2 is recognized by Hsc70 and degraded via chaperone-mediated autophagy to facilitate LD mobilization
To determine how Plin2-mediated lipid hydrolysis is regulated during exit from pluripotency, we performed immunoprecipitation-mass spectrometry to identify proteins interacting with Plin2. The MS analysis revealed that several heat shock proteins interacted with Plin2 (Supplementary Fig. 3a, b). Among these proteins identified, Hspa8, alternately known as Hsc70, is a key regulator in CMA, which targets CMA substrate proteins to lysosome [33]. Because CMA activity is upregulated during differentiation of mESCs [34], and Plin2 is a substrate of CMA [35], we hypothesized that Plin2 is degraded by CMA to facilitate lipid hydrolysis upon exit from pluripotency. We examined the interaction of Plin2 and Hsc70 in undifferentiated and differentiating mESCs. Co-IP using Hsc70 antibody showed the binding of Plin2 to Hsc70 increased remarkably on day 3 of differentiation (Fig. 3a). Immunofluorescence analysis revealed the co-localization of Plin2-GFP with Hsc70 increased significantly on day 3 of differentiation (Fig. 3b), suggesting the binding of Plin2 to Hsc70 depends on differentiation. We also included IDH1 as positive control and another LD protein Abhydrolase Domain Containing 5 (ABHD5) as negative control. ABHD5 did not interact with Hsc70 and remained unchanged during differentiation (Supplementary Fig. 3c, d), indicating Plin2 may be specifically degraded during pluripotency exit. We then knocked down lysosome-associated membrane protein type 2A (LAMP2A), the rate-limiting component of CMA [36], to suppress CMA during differentiation. Plin2 and IDH1 protein levels were increased markedly, while ABHD5 remained unchanged (Fig. 3c and Supplementary Fig. 3e), indicating Plin2 is degraded by CMA upon pluripotency exit of mESCs. Inhibition of CMA by LAMP2A knockdown decreased lipid hydrolysis and suppressed the differentiation of mESCs (Fig. 3d, e), whereas activation of CMA using QX77 increased lipid hydrolysis and promoted pluripotency exit (Fig. 3f, g). Furthermore, activation of CMA by QX77 enhanced the interaction of Plin2 with Hsc70 and decreased Plin2 protein level (Supplementary Fig. 3f, g). These results suggest a potential role for CMA-mediated Plin2 degradation in the early differentiation of mESCs.
a CO-IP of Plin2 and Hsc70 in undifferentiated (day 0) and differentiating mESCs (day 3). The experiments were repeated independently twice with similar results. b Representative images and quantification of the co-localization of Plin2-GFP with Hsc70 in undifferentiated (day 0) and differentiating mESCs (day 3). Scale bar, 5 μm. Data are mean ± SD, n = 3 independent experiments from 30 cells. Two-tailed unpaired t-tests. ***P < 0.001. c Western blot analysis of Plin2 in differentiating mESCs (day 3) transfected with control vector (shCT) or two different shRNAs against LAMP2A. The experiments were repeated independently twice with similar results. d Remaining TG content in differentiating mESCs (day 3) transfected with shCT or shRNAs against LAMP2A. Remaining TG is expressed as a percentage of the TG content at 0 h in the hydrolysis assay. Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. **P < 0.005, *P < 0.05. e Quantification of Oct4-GFP+ cells by flow cytometry in mESC transfected with shCT or shRNAs against LAMP2A on day 3 of differentiation. Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. ***P < 0.001, *P < 0.05. f Remaining TG content in differentiating mESCs (day 3) treated with DMSO or QX77. Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. *P < 0.05. g Quantification of Oct4-GFP+ cells by flow cytometry in mESC treated with DMSO or QX77 on day 3 of differentiation. Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. *P < 0.05. h Schematic depicting the construct of wild-type (WT) and CMA-resistant (MT) Plin2 and representative images and quantification of the co-localization of Plin2-GFP with Hsc70 in differentiating mESCs (day 3) transfected with WT-Plin2-GFP or MT-Plin2-GFP. Scale bar, 5 μm. Data are mean ± SD, n = 3 independent experiments from 30 cells. Two-tailed unpaired t-tests. ***P < 0.001. i Remaining TG content in WT and Plin2-/- mESCs (KO1 and KO2) transfected with empty vector (EV) or MT-Plin2 on day 3 of differentiation. Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. ***P < 0.001, **P < 0.005, *P < 0.05. j Quantification of Oct4-GFP+ cells by flow cytometry in mESC transfected with EV, WT-Plin2 or MT-Plin2 on day 3 of differentiation. Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. **P < 0.005, *P < 0.05. k qRT-PCR analysis of expression of pluripotency markers (Oct4, Nanog, Rex1) in WT and Plin2-/- mESCs (KO1 and KO2) transfected with EV or MT-Plin2 on day 3 of differentiation. Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. ***P < 0.001, **P < 0.005.
To further examine whether CMA-mediated degradation of Plin2 contributes to lipid hydrolysis during pluripotency exit of mESCs, we mutated the CMA targeting motif of Plin2 and measured its co-localization with Hsc70 (Fig. 3h). CMA-resistant Plin2 (MT-Plin2) exhibited decreased co-localization with Hsc70 on day 3 of differentiation (Fig. 3h). WT-Plin2 was degraded on day 3 of differentiation, while MT-Plin2 remained largely unchanged (Supplementary Fig. 3h). These results demonstrate that MT-Plin2 loses its binding to Hsc70 and resists degradation during early differentiation. Expression of CMA-resistant Plin2 (MT-Plin2) resulted in markedly decreased lipid hydrolysis, as shown by the higher levels of remaining TG after Triacsin C treatment (Fig. 3i), suggesting a substantial contribution of CMA-mediated Plin2 degradation to increased lipid hydrolysis during pluripotency exit of mESCs. Flow cytometry analysis showed that expression of WT-Plin2 retarded mESC exit from pluripotency, while MT-Plin2 delayed pluripotency exit more efficiently (Fig. 3j). Moreover, MT-Plin2 significantly restored the expression of pluripotency genes in Plin2-/- mESCs (Fig. 3k). Collectively, these results show that CMA-mediated degradation of Plin2 facilitates lipid hydrolysis and accelerates ESC exit from pluripotency.
Loss of Plin2 induces lipidomic remodeling associated with impaired mitochondrial cristae and FAO
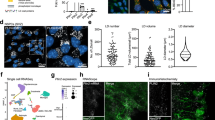
The storage and mobilization of lipids play a crucial role in cellular lipid homeostasis [22]. To investigate the changes in cellular lipid composition upon Plin2 deficiency in mESCs, we conducted nontargeted lipidomics on WT and Plin2-/- mESCs. A total of 1076 distinct lipid molecules were identified and quantified (Supplementary Table 4). Principal component analysis of lipidomic data revealed that KO1 and KO2 mESCs were tightly clustered while WT mESCs were grouped as a distinct population, demonstrating that the major changes in lipids was dependent on Plin2 (Fig. 4a). We found that most lipid species decreased significantly in Plin2-/- mESCs, and only few increased, indicating that Plin2 deficiency caused a significant change in the lipid profiles of mESCs (Fig. 4b). The most prominent changes involved decreases in cardiolipin (CL) and phosphatidylethanolamine (PE) species, and an increase in phosphatidylcholines (PC) (Fig. 4b). We found, particularly that the levels of most CL species identified were lower in Plin2-/- mESCs compared to WT mESCs (Supplementary Fig. 4a). The changes in total level of CL, PE and PC were further verified by enzymatic assays (Supplementary Fig. 4b). These data suggest that loss of Plin2 induces a lipidomic remodeling characterized by decreased CL and PE, and increased PC.
a Principal component analysis (PCA) of lipidomic data from WT and Plin2-/- mESCs (KO1 and KO2). Each dot represents a biological replicate. b Volcano plot of differential lipid molecules between WT and Plin2-/- mESCs (KO1 and KO2). Dot size reports P value, while the color indicates fold change (FC). Lipid molecules with P < 0.05 and FC > 1.5 are considered statistically significant. The top 10 lipid molecules with FC > 2.5 are indicated. c CL, PE, and PC contents in mitochondria of WT and Plin2-/- mESCs (KO1 and KO2). Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. ***P < 0.001, **P < 0.005, *P < 0.05. d Oxygen consumption rates (OCR) and maximal mitochondrial respiration in WT and Plin2-/- mESCs (KO1 and KO2). Maximal mitochondrial respiration was determined by the increase of OCR after FCCP treatment. Data are mean ± SEM, n = 2 biological replicates with three technical replicates each. Two-tailed unpaired t-tests. ***P < 0.001. e Endogenous fatty acid oxidation (FAO) rates and maximal endogenous FAO in WT and Plin2-/- mESCs (KO1 and KO2). Maximal endogenous FAO was determined by the increase of OCR after Eto treatment in a substrate-limited assay medium. Data are mean ± SEM, n = 3 biological replicates with three technical replicates each. Two-tailed unpaired t-tests. ***P < 0.001, *P < 0.05. f Exogenous FAO rates and maximal exogenous FAO in WT and Plin2-/- mESCs (KO1 and KO2). Maximal exogenous FAO was determined by the increase of OCR after PA treatment in a substrate-limited assay medium. Data are mean ± SEM, n = 3 biological replicates with three technical replicates each. Two-tailed unpaired t-tests. ***P < 0.001. g Representative TEM images of mitochondria and quantification of mitochondria with disorganized cristae in WT and Plin2-/- mESCs (KO1 and KO2). Representative mitochondria with normal or disorganized cristae are shown below. Scale bar, top: 1 μm, below: 200 nm. Data are mean ± SD, n = 3 biological replicates (each >50 mitochondria). Two-tailed unpaired t-tests. **P < 0.005, *P < 0.05. h Pie charts of mitochondrial proteomic data showing proportion of mitochondrial proteins changed significantly (P < 0.05 and FC > 1.5) in Plin2-/- mESCs (KO1 and KO2) compared with WT mESCs. Mitochondrial proteins are divided into four categories according to their distribution (inner membrane, outer membrane, matrix or intermembrane space). The experiments were repeated twice with similar results, and one replicate was used for this analysis. i Quantification of mitochondria with disorganized cristae in WT and Plin2-/- mESCs (KO1 and KO2) transfected with EV or MT-Plin2. Data are mean ± SD, n = 3 biological replicates (each >50 mitochondria). Two-tailed unpaired t-tests. **P < 0.005. j Maximal mitochondrial respiration in WT and Plin2-/- mESCs (KO1 and KO2) transfected with EV or MT-Plin2. Data are mean ± SEM, n = 3 independent experiments. Two-tailed unpaired t-tests. ***P < 0.001, **P < 0.005.
Because CL and PE constitute approximately 45–60% of mitochondrial phospholipids and are critical for maintaining mitochondrial structure and functions [37], we measured phospholipid content in isolated mitochondria and observed a similar change in mitochondrial CL, PE and PC (Fig. 4c and Supplementary Fig. 4c). We then measured mitochondrial respiration and glycolysis to assess cellular bioenergetics. The real-time analysis of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) showed that Plin2-/- mESCs exhibited significantly decreased OCR, but had similar ECAR as compared to WT mESCs, suggesting impaired mitochondrial respiration in Plin2-/- mESCs (Fig. 4d and Supplementary Fig. 4d). Fatty acids (FAs) are important substrates for mitochondrial respiration and closely related to LDs. Therefore, we asked whether FAO contributes to decreased OCR in Plin2-/- mESCs. The ability of oxidation and utilization of endogenous FAs were determined by the increase of OCR after treatment of etomoxir (Eto), an inhibitor of mitochondrial FAO, in a substrate-limited assay medium, while oxidation of exogenous FAs were determined by using palmitate (PA)-BSA substrate. Inhibition of FAO by Eto caused considerably more decrease in maximal OCR in WT mESCs than in Plin2-/- mESCs (Fig. 4e), indicating a large portion of OCR decreased is due to oxidation of endogenous FAs. Furthermore, WT mESCs showed significantly increased maximal respiration when treated with exogenous PA, while Plin2-/- mESCs has relatively smaller change in maximal respiration (Fig. 4f), which indicate lower FAO capacity in Plin2-/- mESCs. These results suggest that Plin2 deficiency leads to mitochondrial dysfunction associated with decreased FAO capacity.
Total mitochondrial respiration is closely linked to the number, morphology and structure of mitochondria. Although Plin2-/- mESCs maintained mitochondrial number and morphology similar to WT mESCs (Supplementary Fig. 4e–g), the mitochondrial cristae structure in Plin2-/- mESCs was strikingly different from that in WT mESCs (Fig. 4g). Plin2-/- mESCs exhibited disorganized mitochondrial inner membrane, suggesting loss of mitochondrial cristae structure upon Plin2 knockout (Fig. 4g). The structural integrity of cristae is primarily maintained by the cooperation of lipids and proteins in the mitochondrial inner membrane [38]. We next analyzed mitochondrial protein content by quantitative proteomics on isolated mitochondria, from which we identified 524 mitochondrial proteins (Fig. 4h). Most of proteins of the mitochondrial inner membrane (50.49%, 104/206), intermembrane space (54.55%, 24/44) and matrix (63.64%, 119/187) were significantly decreased in Plin2-/- mESCs, while the majority of proteins in mitochondrial outer membrane remained unchanged (Fig. 4h). The decrease of proteins in mitochondrial inner membrane is in line with the impaired mitochondrial cristae. Importantly, expression of MT-Plin2 recovered mitochondrial cristae and maximal respiration in Plin2-/- mESCs (Fig. 4i, j). Altogether, Plin2 deficiency leads to lipidomic remodeling associated with loss of mitochondrial cristae structure and impaired FAO, indicating an unexpected role for mobilization of LDs in maintaining the structure and function of mitochondria in mESCs.
Loss of Plin2 reduces acetyl-CoA and histone acetylation in mESCs
To better understand how loss of Plin2 and LDs impacts cellular metabolic state and pluripotency, we analyzed key metabolites of glycolysis, the pentose phosphate pathway, the TCA cycle, and nucleic acid metabolism by liquid chromatography-mass spectrometry (LC-MS) and gas chromatography-mass spectrometry (GC-MS) in WT mESCs and Plin2-/- mESCs. Almost all the tested metabolites decreased markedly in mESCs after 3 days of differentiation (Fig. 5a). Notably, undifferentiated Plin2-/- mESCs exhibited significantly lower levels of acetyl-CoA, citrate and NAD+ compared to WT (Fig. 5a), all of which are important intermediates in cellular bioenergetics and are closely linked to mitochondrial activity [39, 40]. Because activation of CMA by QX77 decreased cellular acetyl-CoA levels (Supplementary Fig. 5a), we asked whether acetyl-CoA derived from FAO contributes to pluripotency maintaining of mESCs. After confirmation of decreased acetyl-CoA by enzymatic assay (Fig. 5b), we tested the effects of citrate on acetyl-CoA and pluripotency. Citrate supplementation increased cellular acetyl-CoA levels and rescued pluripotency in Plin2-/- mESCs (Supplementary Fig. 5b, c), suggesting acetyl-CoA may be responsible for pluripotency exit of Plin2-/- mESCs. We further examined the contribution of FA-derived carbons to acetyl-CoA by incubating with [U-13C16] palmitic acid. The incorporation of [U-13C16] palmitic acid in acetyl-CoA was decreased markedly in Plin2-/- mESCs compared with WT mESCs (Fig. 5c), indicating that large amounts of acetyl-CoA are derived from FAO in mESCs.
a Heatmap showing the relative levels of metabolites involved in glycolysis, the pentose phosphate pathway, the citric acid cycle, and nucleotides in WT and Plin2-/- mESCs (KO1 and KO2) on day 0 and day 3 of differentiation. Data are represented as z-score calculated from mean of three biological replicates. b Acetyl-CoA content in WT and Plin2-/- mESCs (KO1 and KO2) on day 0 and day 3 of differentiation. Data are represented as mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. **P < 0.005. c The fraction of M+2 acetyl-CoA in WT and Plin2-/- mESCs (KO1 and KO2) cultured with [U-13C16] palmitic acid for 24 h. Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. **P < 0.005. d Western blot analysis of histone acetylation and quantification of relative H3K27ac levels in WT and Plin2-/- mESCs (KO1 and KO2) on day 3 of differentiation. Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. **P < 0.005. e Normalized signal intensity of H3K27ac-associated genes from ChIP-seq data of WT and Plin2-/- mESCs (KO1 and KO2) on day 3 of differentiation. The experiments were repeated independently twice with similar results, and one biological replicate was used for analysis. f Pie charts of ChIP-seq data showing the proportions of pluripotency genes (112) and differentiation genes (167) with decreased H3K27ac levels (green portion) and unchanged or increased H3K27ac (blue portion) in WT and Plin2-/- mESCs (KO1 and KO2) on day 3 of differentiation. The experiments were repeated independently twice with similar results, and one biological replicate was used for analysis. g Normalized signal intensity of H3K27ac on pluripotency genes from ChIP-seq data of WT and Plin2-/- mESCs (KO1 and KO2) on day 3 of differentiation. The experiments were repeated independently twice with similar results, and one biological replicate was used for analysis. h H3K27ac enrichment around selected pluripotency genes (Oct4, Nanog, and Rex1) from ChIP-seq data of WT and Plin2-/- mESCs (KO1 and KO2) on day 3 of differentiation. The experiments were repeated twice with similar results, and one biological replicate was used for analysis. i Acetyl-CoA content in WT and Plin2-/- mESCs (KO1 and KO2) transfected with EV or MT-Plin2. Data are represented as mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. ***P < 0.001. j Western blot analysis and quantification of relative H3K27ac levels in WT and Plin2-/- mESCs (KO1 and KO2) transfected with EV or MT-Plin2 on day 3 of differentiation. Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. **P < 0.005, *P < 0.05. k ChIP-qPCR analysis of H3K27ac enrichment on promoter regions of selected pluripotency genes (Oct4, Nanog, and Rex1) in WT and Plin2-/- mESCs (KO1 and KO2) transfected with EV or MT-Plin2 on day 3 of differentiation. Data are mean ± SD, n = 2 independent experiments from two primers. Two-tailed unpaired t-tests. ***P < 0.001, **P < 0.005, *P < 0.05.
Given the central roles of acetyl-CoA in regulating histone acetylation [39], we asked whether histone acetylation is responsible for the early differentiation of Plin2-/- mESCs. We examined global acetylation status of histones H3 and H4 during early differentiation of WT mESCs and Plin2-/- mESCs by western blot. The H3K27ac was markedly reduced in Plin2-/- mESCs compared with WT mESCs (Fig. 5d). To gain deeper insights into the changes in histone acetylation, we carried out chromatin immunoprecipitation sequencing (ChIP-seq) to measure genome-wide changes in H3K27ac. Genome-wide ChIP-seq analyses showed the H3K27ac intensities around transcriptional start sites (TSS) were significantly decreased in Plin2-/- mESCs (Fig. 5e and Supplementary Fig. 5d). We then analyzed changes in H3K27ac at promoter regions of pluripotency genes and differentiation-related genes. Over half of pluripotency genes showed decreased H3K27ac in Plin2-/- relative to WT mESCs, while only a small number of genes related to differentiation had decreased H3K27ac (Fig. 5f). The H3K27ac intensities around TSS of pluripotency genes were also significantly reduced in Plin2-/- mESCs (Fig. 5g and Supplementary Fig. 5d). Importantly, decreased H3K27ac was observed at key pluripotency genes, including Oct4, Nanog, and Rex1 (Fig. 5h). Expression of MT-Plin2 restored cellular acetyl-CoA and global H3K27ac in Plin2-/- mESCs (Fig. 5i, j). Furthermore, MT-Plin2 expression increased H3K27ac on promoter regions of key pluripotency genes (Fig. 5k). These results suggest that loss of LDs results in a selective decrease in H3K27ac levels at pluripotency-associated genes during early differentiation of mESCs.
Histone acetyltransferases (HATs) and deacetylases (HDACs) are important for the regulation of histone acetylation and pluripotency [41, 42]. To examine whether the contribution of HATs and HDACs to Plin2-mediated pluripotency, we analyzed the expression of HATs and HDACs, including Hdac1, 2, 3, 6 and Kat2, 3b, 5, 7, 8, all of which have been shown to play a role in pluripotency regulation [41, 42] (Supplementary Fig. 6a, b). No significant differences were observed between WT and Plin2-/- mESCs (Supplementary Fig. 6a, b). We then measured total HAT and HDAC activity in WT and Plin2-/- mESCs. The total activity of HATs was decreased in Plin2-/- mESCs, while the activity of HDACs remained unchanged (Supplementary Fig. 6c, d), which may be due to the reduced cellular acetyl-CoA. Among these HATs, P300 (Kat3b) is the main HAT that mediates H3K27ac in mESCs [43]. To test whether P300 is responsible for the selective decreased H3K27ac at pluripotency genes, we suppressed P300 by a selective catalytic P300 inhibitor C646. Although MT-Plin2 expression increased the expression of pluripotency genes in Plin2-/- mESCs, it could not recover pluripotency gene expression in the presence of C646, suggesting that P300 is critical for Plin2-mediated pluripotency (Supplementary Fig. 6e). We also performed ChIP-qPCR to analyze P300 enrichment around promoters of pluripotency genes and genes of the three germ layers. Promoters of pluripotency genes had greater enrichment of P300 than germ layer marker genes, suggesting pluripotency genes may be more sensitive to acetyl-CoA levels (Supplementary Fig. 6f). We also observed a reduction of P300 binding at pluripotency genes in Plin2-/- mESCs relative to WT (Supplementary Fig. 6f). Therefore, selectively decreased H3K27ac at pluripotency genes in Plin2-/- mESCs may be largely due to the differential P300 distribution between genes associated with pluripotency and differentiation. Altogether, these data demonstrate that Plin2 deficiency reduces acetyl-CoA and H3K27ac to accelerate pluripotency exit, indicating the interplay between LD mobilization and mitochondria-related metabolites in epigenetic regulation.
Enhanced lipid hydrolysis is responsible for phospholipid remodeling and mitochondrial defects in Plin2 -/- mESCs
To clarify the causal relationship among Plin2-mediated lipid hydrolysis, lipidomic remodeling, and histone acetylation in mESC pluripotency, we asked whether inhibition of lipid hydrolysis could reverse the lipid profiles and associated mitochondrial defects in Plin2-/- mESCs. Inhibition of lipid hydrolysis by ATGLi increased mitochondrial CL and PE levels, whereas decreased mitochondrial PC levels in Plin2-/- mESCs (Fig. 6a), suggesting Plin2 deficiency causes mitochondrial phospholipid remodeling by increasing lipid hydrolysis. To further access the effects of lipid hydrolysis-driven phospholipid remodeling on mitochondrial defects, we analyzed mitochondrial cristae and respiration in WT and Plin2-/- mESCs in the presence of ATGLi. Notably, ATGLi treatment partially recovered disorganized mitochondrial cristae and increased mitochondrial OCR in Plin2-/- mESCs (Fig. 6b, c). Moreover, total acetyl-CoA and H3K27ac levels in Plin2-/- mESCs were increased by ATGLi treatment (Fig. 6d, e). These results suggest that enhanced lipid hydrolysis is responsible for the defects in mitochondrial cristae and respiration, which are crucial for maintaining acetyl-CoA and histone acetylation in Plin2-/- mESCs.
a CL, PE, and PC contents in mitochondria of WT and Plin2-/- mESCs (KO1 and KO2) treated with DMSO or ATGLi (10 μM Atglistatin). Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. ***P < 0.001, **P < 0.005. b Representative TEM images of mitochondria and quantification of mitochondria with disorganized cristae in WT and Plin2-/- mESCs (KO1 and KO2) treated with DMSO or ATGLi (10 μM Atglistatin). Scale bar, 1 μm. Data are mean ± SD, n = 3 biological replicates (each >100 mitochondria). Two-tailed unpaired t-tests. **P < 0.005. c Maximal mitochondrial respiration in WT and Plin2-/- mESCs (KO1 and KO2) treated with DMSO or ATGLi (10 μM Atglistatin). Data are mean ± SEM, n = 3 biological replicates. Two-tailed unpaired t-tests. ***P < 0.001. d Acetyl-CoA content in WT and Plin2-/- mESCs (KO1 and KO2) treated with DMSO or ATGLi (10 μM Atglistatin). Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. *P < 0.05. e Western blot analysis and quantification of H3K27ac levels in WT and Plin2-/- mESCs (KO1 and KO2) treated with DMSO or ATGLi (10 μM Atglistatin) on day 3 of differentiation. Data are mean ± SD, n = 3 biological replicates. Two-tailed unpaired t-tests. ***P < 0.001.
To gain insights into the functional role of specific phospholipid species in mitochondrial function and pluripotency regulation, we supplemented WT and Plin2-/- mESCs with Lyso-phosphatidylethanolamine (Lyso-PE), which has been shown to preferentially be converted to mitochondrial PE [44], and measured the expression of pluripotency genes during early differentiation. Supplementation with Lyso-PE delayed the loss of pluripotency genes in Plin2-/- mESCs, but had no effect in WT mESCs, as shown by the expression of Oct4 and Nanog (Supplementary Fig. 7a). In addition, Lyso-PE supplementation increased mitochondrial PE levels in both WT and Plin2-/- mESCs, without altering mitochondrial CL levels (Supplementary Fig. 7b, c), suggesting mitochondrial PE retards pluripotency exit in a CL-independent manner. Because mitochondrial activity has been suggested to play a role in PSCs [14, 45], and mitochondrial PE could enhance mitochondrial respiration by multiple mechanisms [44, 46], we asked whether supplementation with Lyso-PE could recover defects in mitochondrial respiration. Notably, Lyso-PE supplementation resulted in a significant increase in OCR of Plin2-/- mESCs, while a relatively small increase in that of WT mESCs (Supplementary Fig. 7d). Similar results were observed in acetyl-CoA and H3K27ac levels (Supplementary Fig. 7e, f), which is consistent with the results of increased pluripotency genes. Collectively, these results demonstrate that mitochondrial PE restores pluripotency gene expression in Plin2-/- mESCs by improving mitochondrial respiration, indicating that impaired mitochondrial activity may be responsible for the pluripotency exit caused by excessive lipid hydrolysis.
Discussion
Metabolic reprogramming plays crucial roles in cell fate determination. However, both the regulation and functions of lipid metabolism in cell fate decision of PSCs remain unclear, especially with regard to lipid hydrolysis. In the present study, we show that Plin2-mediated LD mobilization is critical for pluripotency of mESCs by coordinating phospholipid homeostasis, mitochondrial activity, and histone acetylation. Upon exit from pluripotency, Plin2 is degraded by CMA to facilitate ATGL-mediated lipid hydrolysis. Loss of Plin2 causes a dramatic lipidomic remodeling associated with disorganized mitochondrial cristae, leading to mitochondrial respiration defects that decrease cellular acetyl-CoA content and histone acetylation, ultimately making mESC prone to exit from pluripotency (Fig. 7). These results reveal a mechanism by which mitochondrial remodeling links lipidome with epigenome to control stem cell fate.
Plin2-mediated LD mobilization is critical for pluripotency of mESCs by coordinating phospholipid homeostasis, mitochondrial activity, and histone acetylation. LD-associated protein Plin2 protects LD from ATGL to maintain low lipid hydrolysis in mESCs. Upon exit from pluripotency, Plin2 is recognized by Hsc70 and degraded by chaperone-mediated autophagy to facilitate ATGL-mediated lipid hydrolysis. Excessive lipid hydrolysis induces a dramatic lipidomic remodeling characterized by decreased cardiolipin and phosphatidylethanolamine, which triggers defects in mitochondrial cristae and fatty acid oxidation. Impaired fatty acid oxidation results in decreased cellular acetyl-CoA content and histone acetylation, which are crucial for the expression of pluripotency genes, ultimately making mESC prone to exit from pluripotency.
Our study reveals a critical role for LD homeostasis in pluripotency maintenance and elucidates how LD mobilization is regulated during pluripotency exit of mESCs. LDs are central organelles controlling lipid homeostasis by coordinating lipid storage and hydrolysis. Though considerable numbers of LDs were observed in mammal oocytes and embryos and cultured PSCs [26], whether and how LD homeostasis participates in embryonic development and pluripotency regulation remain largely unknown. Recent studies revealed that a proper level of LDs is required for early embryonic development using forced lipophagy- or delipidation-based methods to remove LDs, and increased LD mobilization is critical for maintaining embryonic diapause [20, 21, 47]. In addition, LD accumulation and increased lipogenesis were observed during the process of pluripotency acquisition and the transition from a naive pluripotent state to a primed state [16, 27], suggesting important roles for LD homeostasis in PSCs. However, how LD homeostasis is regulated and its functional roles in PSCs are still obscure. We show lipid hydrolysis is increased via CMA during pluripotency exit of mESCs, which is in agreement with recent findings showing CMA activation promotes mESC differentiation [34]. As posttranslational modifications of Plin2 is critical for its CMA degradation [48], it will be interesting to clarify how posttranslational modifications of Plin2 regulate its interaction with Hsc70 and subsequent CMA degradation. Our findings extend current knowledge about how CMA orchestrates mESC pluripotency, and identify Plin2 as the critical LD-associated protein that suppresses LD mobilization to facilitate pluripotency maintaining.
Our data suggest important roles for LD homeostasis and mitochondria activity in epigenetic control of pluripotency gene expression. Mitochondrial cristae integrity is regulated by phospholipid homeostasis, which is linked to histone acetylation via FAO and acetyl-CoA, suggesting the importance of mitochondrial FAO in maintaining acetyl-CoA pools in PSCs. Recently, it has been reported that FAO plays a conserved role in restricting the quiescence of naive PSCs [49]. Our observations are broadly consistent with previous reports of bivalent metabolic state in naive PSCs and provide insights into the potential roles of FAO in pluripotency maintaining [16, 50]. In addition, increased lipid hydrolysis, impaired mitochondrial activity, phospholipid remodeling, and reduced histone acetylation have been observed in embryonic diapause and diapause-like state [47, 49, 51]. However, the mechanism linking these events is unclear. Our results uncover the detailed mechanism of the interplay among LD mobilization, lipidomic remodeling, mitochondrial activity, and epigenetic alterations, and highlight the regulation of mitochondrial cristae by lipid homeostasis, which may provide insights into embryonic diapause and potential therapeutic opportunities for reproductive health. Finally, as mitochondrial structure and LDs are strikingly different between the naive and primed pluripotent states [16], our data suggest a potential association between LD homeostasis and mitochondrial integrity in the transitions between different pluripotent states, which merit further investigation.
ATGL has been reported to enhance mitochondrial function in many metabolically active tissues [52], which is inconsistent with our observation that inhibition of ATGL restores mitochondrial respiration function. Although defective lipolysis is often associated with impaired mitochondrial function, increased lipolysis and impaired mitochondria have been observed in several cell types, including diapause embryos, quiescent ESCs, and regulatory T cells [47, 51,52,53], implying the interplay between lipolysis and mitochondrial function is context-dependent. The discrepancy in ATGL function may be due to the striking differences in metabolic state and mitochondrial function between somatic cells and ESCs. In addition, the mechanism by which lipid hydrolysis regulates mitochondrial phospholipid remodeling remains unclear. Considering mitochondrial CL and PE could be synthesized by mitochondrial enzymes, whereas mitochondrial PC needs to be imported from other organelles [54], possible mechanisms include but are not limited to the differences in substrate utilization, enzyme expression, lipid transfer, and so on. Further studies will be necessary to elucidate the interplay between lipid hydrolysis and phospholipid homeostasis.
In conclusion, we demonstrate that Plin2 safeguards pluripotency of mESCs by suppressing LD mobilization, and CMA-mediated degradation of Plin2 facilitates lipid hydrolysis to drive the pluripotency exit of mESCs. Mechanistically, excessive LD mobilization results in phospholipid remodeling and disorganized mitochondrial cristae, thereby reducing mitochondrial FAO and acetyl-CoA, which is crucial for maintaining histone acetylation. Our results reveal the mechanism by which LD mobilization regulates pluripotency, and provide insights into the interplay between LD homeostasis and mitochondrial remodeling. Given the increasing number of pathological conditions associated with LD accumulation and mitochondrial remodeling, such as cancer progression and neural degeneration, our findings will shed light on the mechanisms of these pathological processes.
Methods
Cell culture
OG2 mESCs and OG2 MEFs were derived from E3.5 and E13.5 embryos, respectively, carrying the Oct4-GFP transgenic allele. R1 mESCs were bought from ATCC. OG2 and R1 mESCs were cultured on 0.1% gelatin-coated plates in DMEM/high glucose supplemented with N2 (Gibco, 200×), B27 (Gibco, 100×), sodium pyruvate (Gibco, 100×), GlutaMAX (Gibco, 100×), NEAA (Gibco, 100×), 0.1 mM 2-mercaptoethanol (Gibco), 1000 U/mL leukemia inhibitory factor (LIF), 3 μM CHIR99021 (Selleck) and 1 μM PD0325901 (Selleck). OG2 MEFs, Plat-E, and 293T cells were maintained in DMEM/high glucose (Hyclone) supplemented with 10% FBS (Gibco), GlutaMAX (Gibco, 100×), and NEAA (Gibco, 100×). Atglistatin was used at 10 μM. Dox was used at 1 μg/mL. The cells were obtained with approval from the ethics committee of the Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences (GIBH). All the animals were handled according to approved Institutional Animal Care and Use Committee protocols of the GIBH.
Plasmids
pMXs retroviral vector expressing Sox2, Klf4, Oct4, or c-Myc and pX330 vector were purchased from Addgene. shRNA inserts were cloned into pRetroSuper retroviral vector or pLKO.1 lentiviral vector. The coding sequence of Plin2, HA-tagged Plin2, and CMA-resistant Plin2 were cloned into pLenti-puro or pCW-puro lentiviral vector. The shRNA sequences used are listed in Supplementary Table 1.
Reprogramming of MEFs to iPSCs
Retroviral vectors expressing mouse Sox2, Klf4, Oct4, and c-Myc (SKOM) were transfected in Plat-E cells using polyethylenimine (PolyScience) to produce retrovirus. A total of 1.5 × 104 OG2 MEFs within two passages were seeded in 12-well plates and infected twice with retroviral supernatants generated with Plat-E cells. After infection, cells were cultured in DMEM/high glucose (Hyclone) supplemented with 15% FBS (Gibco), sodium pyruvate (Gibco, 100×), GlutaMAX (Gibco, 100×), NEAA (Gibco, 100×), 0.1 mM 2-mercaptoethanol (Gibco), 1000 U per mL LIF. Oct4-GFP positive colonies were counted on day 14 after infection.
Conversion of naive mESCs to primed mESCs
Naive mESCs were seeded on serum-coated 6-well plates at a density of 3 × 104 cells per well and cultured in FN medium for 5 days. The FN medium comprised 50% DMEM/F12 (Gibco), 50% Neurobasal (Gibco), N2 (Gibco, 200×), B27 (Gibco, 100×), GlutaMAX (Gibco, 100×), NEAA (Gibco, 100×), 0.1 mM 2-mercaptoethanol (Gibco), 1 mg/mL BSA (Sigma), 20 ng/mL Activin A (PeproTech), 20 ng/mL bFGF (PeproTech), 2 μM XAV939 (Selleck), and 3 μM CHIR99021 (Selleck).
Spontaneous differentiation of mESCs
To induce spontaneous differentiation, mESCs were dissociated into single cells using 0.25% trypsin-EDTA (Gibco). The cell suspension (6 × 105 cells/mL) was plated in 100 mm non-adherent dish and cultured in EB medium for 3 days. The EB medium comprised Knockout DMEM (Gibco), 15% KSR (Gibco), GlutaMAX (Gibco, 100×), and NEAA (Gibco, 100×). After 3 days of EB formation, EBs were plated in a gelatin-coated dish and cultured for indicated days.
Generation of Plin2 knockout mESCs
For generating CRISPR-Cas9-mediated Plin2 knockout in mESCs, guide RNAs (gRNAs) for Plin2 were designed on the website (crispr.mit.edu) and cloned into pX330 vector. For targeting, a total of 1 × 106 mESCs were electroporated with 3 μg pX300 plasmid containing gRNA for Plin2 and then seeded in gelatin-coated plates. After 48 h, ESC colonies were picked and verified by genomic PCR and western blot.
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA was extracted by TRIzol (Invitrogen) and 5 μg RNA was used to generate complementary DNA. Transcriptional levels of genes were determined by Premix Ex Taq (Takara) or ChamQ SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd) and analyzed with a CFX-96 Real-Time system (Bio-Rad). The primers used are listed in Supplementary Table 2.
Western blot
Whole-cell extracts were obtained with radioimmunoprecipitation (RIPA) buffer (Beyotime) supplemented with protease inhibitor cocktail (Bimake). Nuclear protein extracts were prepared with the kit (Beyotime) according to the manufacturer’s instructions. Proteins were analyzed with SDS-PAGE and then transferred to PVDF membrane (Millipore). After incubation with antibodies, the membrane was exposed to X film. The following primary antibodies were used: anti-GAPDH (Bioworld, AP2063, 1:5000), anti-Plin2 (Proteintech, 15294-1-AP, 1:200 or Abcam, ab108323, 1:500), anti-Oct4 (Cell Signaling, 2750, 1:1000), anti-histone H3 (Abcam, ab1791, 1:5000), anti-H3K4ac (Abcam, ab176799, 1:200), anti-H3K9ac (Abcam, ab4441, 1:200), anti-H3K27ac (Cell Signaling, 8173, 1:1000), anti-H4K16ac (Abcam, ab109463, 1:200), anti-H3Ac (Millipore, 17-615, 1:500), anti-Tom20 (Abcam, ab56783, 1:200), anti-HA (Cell Signaling, 3724, 1:500), anti-LAMP2A (Abcam, ab13524, 1:500), anti-beta-III tubulin (Abcam, ab7751, 1:500), anti-AFP (Proteintech, 14550-1-AP, 1:500), anti-Brachyury (Abcam, ab209665, 1:500), anti-ABHD5 (Proteintech, 12201-1-AP, 1:500), anti-IDH1 (Proteintech, 12332-1-AP, 1:500) and anti-Hsc70 (Santa Cruz, sc7298, 1:1000). The secondary antibodies were goat anti-rabbit (Kangchen, KC-RB-035), goat anti-mouse (Kangchen, KC-MM-035), and goat anti-rat (Abcam, ab182018, 1:2000). Full-length original western blots for these results are provided in Supplementary File 1.
Immunofluorescence
Cells on glass dishes were fixed with 4% paraformaldehyde for 30 min, permeabilized in 0.1% Trition X-100 for 30 min, and then blocked in 10% FBS (Gibco) for 1 h at room temperature. Cells were incubated with primary antibodies for 3 h and then secondary antibodies for 1 h at room temperature. Next, the cells were stained with 0.1 μg/mL DAPI (Sigma) and analyzed by a Zeiss LSM 710 confocal microscope. The primary antibody was anti-Plin2 (Proteintech, 15294-1-AP, 1:50), anti-Hsc70 (Abcam, ab51052, 1:200) or anti-Tom20 (Abcam, ab56783, 1:100). The secondary antibody was goat anti-rabbit DyLight® 594 (Abcam, ab96885, 1:1000).
Flow cytometry
For neutral lipid quantification, cells were washed with PBS and stained with BODIPY 493/503 for 20 min, and then washed with PBS three times and dissociated into single cell with trypsin-EDTA (0.25%). For Oct4-GFP analysis, cells were washed with PBS and dissociated into single cell with trypsin-EDTA (0.25%). The cells were analyzed with an Accuri C6 flow cytometer (BD Biosciences).
Oil Red O staining
Cells on glass dishes were fixed with 4% paraformaldehyde for 30 min, incubated with 60% isopropanol for 5 min, and then stained with 1.8 mg/mL Oil Red O (Sigma) for 5 min at room temperature. Next, the cells were stained with 0.1 μg/mL DAPI (Sigma) and analyzed by a Zeiss LSM 710 confocal microscope.
Lipid hydrolysis assay
Lipid hydrolysis assay was performed as previously described with minor revisions [32]. Culture medium was refreshed 12 h before the assay. Then the culture medium was replaced with lipid-depleted medium (culture medium deprived of B27) containing 5 μM Triacsin C to inhibit TG synthesis from the recycling of newly released FAs. Cells were washed with PBS and TG contents were measured at indicated time points by BODIPY 493/503 or an enzymatic assay. Remaining TG is expressed as a percentage of the TG content at 0 h.
TEM
Cells were washed with PBS and fixed with 3% glutaraldehyde (Sigma) for 12 h at 4 °C. Next, cells were fixed with 1% osmium acid (SPI-Chem) for another 1 h before dehydration by gradient ethanol. After fixation, the samples were stained with lead (SPI-Chem) and uranium (SPI-Chem). After ultra-sectioning with Leica EM UC7, the ultrastructure of LDs and mitochondria was visualized by TEM (FEI Tecnai G2 Spirit, USA) at 120 keV.
Lipid extraction and lipidomics analysis
Lipids were extracted and analyzed as described previously [8]. Briefly, 1 × 107 cells were quenched in 400 μL methanol: deionized water (3:1, v/v) and incubated with 1 mL methyl tert-butyl ether for 1 h at room temperature. Samples were homogenized and incubated with 200 μL deionized water and centrifuged to obtain the upper organic phase containing lipid extracts. Dried lipid films were resuspended in 120 μL acetonitrile (ACN):isopropanol (IPA):water (60:30:5, v/v/v) for MS analysis. The internal standard mixture was added to each kind of sample matrix prior to lipid extraction as described previously [8].
An ACQUITY UHPLC system (Waters, Milford, MA, USA) coupled via an electrospray ion source with a Q Exactive HF MS system (Thermo Fisher Scientific, Rockford, IL, USA) was used for lipid profiling in both full scan and data-dependent MS/MS (ddMS2) modes to obtain lipid information including retention time (tR), accurate masses and/or MS/MS fragments, etc. Data analysis was performed by Dalian ChemDataSolution Information Technology Co. Ltd.
The identification of lipids was achieved by using a commercial database of metabolites called OSI/SMMS2 and the strategy was described in a previous report [55]. To the molecules with the successful matching of MS2, all information of tR, MS1, and MS2 was employed for a more accurate characterization of lipids. After this, LipidSearch (version 4.0) software was further used to validate the identification with a comparison of details of MS2 fragments, including the length of structure chain and position of double bond. The thresholds of parameters of tR, MS1, and MS2 were set to 0.25 min, 0.01 Da, and 0.025 Da in LipidSearch platform, respectively. To the molecules with no matching of MS2, the information of tR and MS1 was employed to annotate the class of lipids, including the length of chain and position of double bond.
OCR and ECAR analysis
OCR and ECAR were analyzed using the XF24 extracellular flux analyzer (Seahorse Biosciences). A total of 1 × 106 cells were plated on gelatin-coated plates overnight at 37 °C. OCR and ECAR were determined with the Seahorse XF Cell Mito Stress Test Kit and Glycolysis Stress Test Kit, respectively. For OCR measurement, 2 μM oligomycin, 1 μM FCCP, 1 μM rotenone, and 1 μM antimycin were used. For ECAR measurement, 10 mM glucose, 2 μM oligomycin, and 100 mM 2-deoxyglucose were used. FAO was determined with the Seahorse XF palmitate-BSA FAO Substrate from the Seahorse XF Palmitate Oxidation Stress Kit according to the manufacturer’s instructions. For FAO measurement, 2 μM oligomycin, 1 μM FCCP, 1 μM rotenone, 1 μM antimycin and 100 μM Etomoxir were used. Maximal endogenous FAO was determined by the increase of OCR after Eto treatment in a substrate-limited assay medium. Maximal exogenous FAO was determined by the increase of OCR after palmitate treatment in a substrate-limited assay medium. The substrate-limited assay medium was derived from oxidation substrates such as glucose, pyruvate, and glutamine.
Isolation of mitochondria
Mitochondria were isolated with the Qproteome Mitochondria Isolation Kit (Qiagen) following the manufacturer’s instruction. In brief, 5 × 106 cells were washed with 0.9% NaCl and lysed in cold lysis buffer from the kit for 10 min at 4 °C. The lysate was centrifuged to correct the supernatant containing cytosolic proteins. The pellet was disrupted in cold disruption buffer from the kit, and then centrifuged to correct the pellet containing nuclei. The supernatant was further centrifuged to obtain the pellet containing mitochondria, and the mitochondrial pellet was washed with mitochondria storage buffer from the kit.
Quantitative proteomic analysis
The mitochondrial pellet was sonicated three times on ice using a high-intensity ultrasonic processor (Scientz) in lysis buffer (8 M urea and protease inhibitor cocktail). The remaining debris was removed by centrifugation at 12,000 g at 4 °C for 10 min. Finally, the supernatant was collected and the protein concentration was determined with BCA kit (Beyotime) according to the manufacturer’s instructions. The mitochondrial extraction was reduced with 5 mM dithiothreitol for 30 min at 56 °C and alkylated with 11 mM iodoacetamide for 15 min at room temperature in darkness. The protein sample was then diluted and digested with trypsin.
The tryptic peptides were dissolved in solvent A (0.1% formic acid in water), directly loaded onto a homemade reversed-phase analytical column (25 cm length, 75 μm i.d.). Peptides were separated with a gradient from 6 to 22% solvent B (0.1% formic acid in acetonitrile) over 43 min, 22 to 30% in 13 min, and climbing to 80% in 2 min then holding at 80% for the last 2 min, all at a constant flow rate of 400 nl/min on a nanoElute UHPLC system (Bruker Daltonics). The peptides were subjected to Capillary source followed by the timsTOF Pro (Bruker Daltonics) mass spectrometry. The electrospray voltage applied was 1.60 kV. Precursors and fragments were analyzed at the TOF detector, with an MS/MS scan range from 100 to 1700 m/z. The timsTOF Pro was operated in parallel accumulation serial fragmentation (PASEF) mode. Precursors with charge states 0–5 were selected for fragmentation, and 10 PASEF-MS/MS scans were acquired per cycle. The dynamic exclusion was set to 30 s.
The resulting MS/MS data were processed using MaxQuant search engine (v.1.6.6.0). Tandem mass spectra were searched against human uniprot database concatenated with reverse decoy database. Trypsin/P was specified as a cleavage enzyme allowing up to four missing cleavages. The mass tolerance for precursor ions was set as 40 ppm in the first search and 40 ppm in the main search, and the mass tolerance for fragment ions was set as 0.04 Da. Carbamidomethyl on Cys was specified as fixed modification, and acetylation on protein N-terminal and oxidation on Met were specified as variable modifications. FDR was adjusted to <1%.
Metabolites analysis
A total of 1 × 106 cells were homogenized in 400 μL methanol: deionized water (3:1, v/v), and centrifuged at 12,000 rpm for 10 min. The supernatant was transferred to a high recovery glass sampling vial (CNW) to vacuum dry at room temperature. The residue was oximated with 35 μL pyridine containing 20 mg/mL methoxyamine hydrochloride (Sigma) at 37 °C overnight and further derivatized with 20 μL N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide at 70 °C for 30 min.
G6P, F6P, FBP, PEP, Pyruvate, L-lactate, R5P, AMP, ATP, GMP, GDP, ADP, cAMP, NAD+, and NADP+ were determined by LC-MS. Metabolomics profiling was performed with a Water UPLC (ACQUITY UPLC® I-Class) system coupled with a Xevo® TQ-XS mass spectrometry (Water Xevo® TQ-XS). Separation of metabolites was achieved in a Water ACQUITY UPLC BEH Amide column (2.1 mm × 100 mm × 1.7 µm). The elution solvents consisted of A (ACN:H2O = 95:5, v/v) and B (ACN:H2O = 5:95, v/v), both containing 10 nM ammonium bicarbonate. The elution gradient was set as follows: 99% A (0.0–0.1 min), 99–30% A (0.1–6 min), 30–99% (6.0–6.5mim), 99–99% A (6.5–10 min). The column temperature was 45 °C and the flow rate was 0.4 mL/min. The data were acquired in both ESI positive and negative modes. The Capillary was 3.0 kV for positive mode and 2.53 kV for negative mode. The Desolvation Temp was 500 °C. The Source Temp was 150 °C. The data were analyzed using the MSD ChemStation G1701EA E.02.02.1431.
Acetyl-CoA, Citrate, cis-Aconitic acid, Isocitrate, alpha-KG, Succinyl-CoA, Succinic acid, Fumarate, and L-Malic acid were determined by GC-MS. Metabolomics profiling was performed with an Aligent 7890A GC coupled with Agilent 5975C mass spectrometer. Separation was achieved on an HP-5ms fused-silica capillary column (30 m × 250 μm i.d.; 5% diphenyl-95% emthylpolysiloxane bonded and crosslinked) with helium as the carrier gas at a constant flow rate of 1 mL/min through the column. The temperatures of front injection, MSD transfer interface, and electron impact (EI) ion source were set to 280, 290, and 230 °C, respectively. The GC oven temperature was set to 100 °C for 3 min, with an increment rate of 10 °C/min to 140 °C, 8 °C/min to 260 °C, 10 °C/min to 310 °C, and a final 5 min maintenance at 310 °C. The EI ionization was 70 eV. After 5 min of solvent delay, the mass data were collected at full scan mode (m/z 50–600). The data were analyzed using the masslynx v4.2.
Palmitic acid incorporation into acetyl-CoA
For determination of palmitic acid incorporation into acetyl-CoA, cells were cultured with [U-13C16] palmitic acid (ISOREAG) for 24 h before metabolite extraction and LC-MS analysis. The UHPLC-MS/MS analysis was performed on an Agilent 1290 Infinity II UHPLC system coupled to a 6470A Triple Quadrupole mass spectrometry (Santa Clara, CA, USA). Samples were injected onto a Waters BEH Amide column (100 mm × 2.1 mm, 1.7 μm) at a flow rate of 0.25 mL/min. The mobile phase consisted of (A) water in 15 mM ammonium acetate with a pH of 9 and (B) 90% acetonitrile. The chromatographic separation was conducted by a gradient elution program as follows: 0–2 min, 95% B; 9 min, 65% B; 10–12 min, 50% B; 12.1–15 min, 95% B. The eluted analysts were ionized in an electrospray ionization source in positive mode (ESI+). The temperatures of ESI+ source drying gas and sheath gas were 300 and 350 °C. The flow rate of ESI+ source drying gas and sheath gas were 5 and 11 L/min, respectively. The pressure of nebulizer was 40 psi, and capillary voltage was 4000 V. The dynamic multiple reaction monitoring was used to acquire data in optimized MRM transition (precursor -> product).
TG, CL, PE, PC, and acetyl-CoA analysis
Cellular and mitochondrial TG, CL, PE, PC, and acetyl-CoA were measured with the triglyceride quantification colorimetric kit (Biovision, K622), cardiolipin assay kit (Biovision, K944), phosphatidylethanolamine assay kit (Biovision, K499), phosphatidylcholine colorimetric kit (Biovision, K576) and PicoProbe acetyl-CoA fluorometric assay kit (Biovision, K317), respectively. For acetyl-CoA measurement, cells were washed with cold PBS and deproteinized using perchloric acid/KOH following the manufacturer’s instruction. The samples were diluted with assay buffer and incubated in CoA quencher for 5 min at room temperature. The fluorescence signal was measured at Excitation/Emission = 535/587 nm. For TG, CL, PE, and PC measurement, cells were washed with cold PBS and extracted with 5% NP-40 according to the manufacturer’s instructions. The signal was measured at OD 570 nm for TG and PC, Excitation/Emission = 535/587 nm for PE, Excitation/Emission = 340/480 nm for CL.
HAT and HDAC activities analysis
Total HAT and HDAC activities were measured with the HAT activity colorimetric assay kit (Biovision, K332) and HDAC activity colorimetric assay kit (Biovision, K331) respectively. Cells were washed with cold PBS and extracted with 5% NP-40 according to the manufacturer’s instructions. The signal was measured at OD 440 nm for HAT activity and OD 405 nm for HDAC activity. HAT and HDAC activities were expressed as the relative OD value per µg protein sample.
ChIP-seq and data analysis
Cells were crosslinked with 1% formaldehyde (Sigma) for 10 min at room temperature and then quenched with 125 mM glycine. Fixed cells were washed three times with cold PBS and centrifuged at 2500 rpm for 5 min. Cell lysis, sonication, and immunoprecipitation were performed with SimpleChIP Enzymatic Chromatin IP Kit (Magnetic Beads) (Cell Signaling) according to the manufacturer’s instructions. Antibodies used for ChIP were anti-H3K27ac (Cell Signaling, 8173, 1:100), anti-P300 (Cell Signaling Technology, 54062, 1:50), IgG (Cell Signaling Technology, 2729, 1:100). ChIP-seq library was prepared following the Illumina ChIP-seq library preparation protocol. Sequencing on a HiSeq 2000 instrument was carried out by Annoroad Gene Technology Co., Ltd (Beijing, China). ChIP-qPCR primers are listed in Supplementary Table 3.
Sequenced reads were filtered by Trimmomatic (0.35), and then mapped to mouse reference genome (mm10) using Bowtie2 (2.2.5). Peaks were called using SICER (1.1.2) with “W200 G200” parameters. Tracks of signal were computed using MACS2 bdgcmp module with parameter “-m ppois”. The signal BigWig files were visualized using computeMatrix, plotHeatmap, and plotProfile module in DeepTools (2.4.2). ChIPpeakAnno (3.16.1) was used for identifying nearby genes from the peaks obtained from MACS and SICER.
Immunoprecipitation
Cells were lysed in cell lysis buffer (50 mM Tris-HCl pH7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, and protease inhibitor cocktail) for 30 min at 4 °C. Anti-HA (Cell Signaling, 3724, 1:200), anti-Hsc70 (Abcam, ab51052, 1:200), or normal rabbit IgG (Cell Signaling, 2729, 1:1000) was linked to Dynabeads with protein A and G (Invitrogen) for 4 h at 4 °C in lysis buffer. Lysates were incubated with the washed antibody-linked Dynabeads overnight at 4 °C. After immunoprecipitation, beads were washed with lysis buffer five times and boiled in SDS buffer for 10 min. The eluents were analyzed by western blot or mass spectrometry.
Statistical analysis
Data are presented as mean ± SD or mean ± SEM. Sample number (n) indicates the number of independent biological samples in each experiment. Statistical comparisons were performed using the unpaired two-tailed Student’s t-tests with GraphPad Prism 8. P value < 0.05 was considered statistically significant.
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary information. Additional data related to this paper may be requested from the authors. ChIP-seq data have been deposited in the Genome Sequence Archive under accession numbers CRA002919 that are publicly accessible at https://bigd.big.ac.cn/gsa.
Code availability
All code needed to evaluate the conclusions in the paper may be requested from the authors.
References
Zhang J, Zhao J, Dahan P, Lu V, Zhang C, Li H, et al. Metabolism in pluripotent stem cells and early mammalian development. Cell Metab. 2018;27:332–8.
Yanes O, Clark J, Wong DM, Patti GJ, Sanchez-Ruiz A, Benton HP, et al. Metabolic oxidation regulates embryonic stem cell differentiation. Nat Chem Biol. 2010;6:411–7.
Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, et al. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–71.
Moussaieff A, Rouleau M, Kitsberg D, Cohen M, Levy G, Barasch D, et al. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 2015;21:392–402.
Dahan P, Lu V, Nguyen RMT, Kennedy SAL, Teitell MA. Metabolism in pluripotency: both driver and passenger? J Biol Chem. 2019;294:5420–9.
Hwang IY, Kwak S, Lee S, Kim H, Lee SE, Kim JH, et al. Psat1-dependent fluctuations in alpha-Ketoglutarate affect the timing of ESC differentiation. Cell Metab. 2016;24:494–501.
TeSlaa T, Chaikovsky AC, Lipchina I, Escobar SL, Hochedlinger K, Huang J, et al. alpha-Ketoglutarate accelerates the initial differentiation of primed human pluripotent stem cells. Cell Metab. 2016;24:485–93.
Wu Y, Chen K, Xing G, Li L, Ma B, Hu Z, et al. Phospholipid remodeling is critical for stem cell pluripotency by facilitating mesenchymal-to-epithelial transition. Sci Adv. 2019;5:eaax7525.
Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–6.
Li L, Chen K, Wang T, Wu Y, Xing G, Chen M, et al. Glis1 facilitates induction of pluripotency via an epigenome-metabolome-epigenome signalling cascade. Nat Metab. 2020;2:882–92.
Guijas C, Montenegro-Burke JR, Warth B, Spilker ME, Siuzdak G. Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat Biotechnol. 2018;36:316–20.
McDonnell E, Crown SB, Fox DB, Kitir B, Ilkayeva OR, Olsen CA, et al. Lipids reprogram metabolism to become a major carbon source for histone acetylation. Cell Rep. 2016;17:1463–72.
Wong BW, Wang X, Zecchin A, Thienpont B, Cornelissen I, Kalucka J, et al. The role of fatty acid beta-oxidation in lymphangiogenesis. Nature. 2017;542:49–54.
Lisowski P, Kannan P, Mlody B, Prigione A. Mitochondria and the dynamic control of stem cell homeostasis. EMBO Rep. 2018;19:e45432.
Zhou W, Choi M, Margineantu D, Margaretha L, Hesson J, Cavanaugh C, et al. HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31:2103–16.
Sperber H, Mathieu J, Wang Y, Ferreccio A, Hesson J, Xu Z, et al. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat Cell Biol. 2015;17:1523–35.
Liu L, Zhang K, Sandoval H, Yamamoto S, Jaiswal M, Sanz E, et al. Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell. 2015;160:177–90.
Wu H, Han Y, Rodriguez Sillke Y, Deng H, Siddiqui S, Treese C, et al. Lipid droplet-dependent fatty acid metabolism controls the immune suppressive phenotype of tumor-associated macrophages. EMBO Mol Med. 2019;11:e10698.
Liu L, MacKenzie KR, Putluri N, Maletic-Savatic M, Bellen HJ. The glia-neuron lactate shuttle and elevated ROS promote lipid synthesis in neurons and lipid droplet accumulation in glia via APOE/D. Cell Metab. 2017;26:719–37 e6.
Tatsumi T, Takayama K, Ishii S, Yamamoto A, Hara T, Minami N, et al. Forced lipophagy reveals that lipid droplets are required for early embryonic development in mouse. Development. 2018;145:dev161893.
Aizawa R, Ibayashi M, Tatsumi T, Yamamoto A, Kokubo T, Miyasaka N, et al. Synthesis and maintenance of lipid droplets are essential for mouse preimplantation embryonic development. Development. 2019;146:dev181925.
Olzmann JA, Carvalho P. Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol. 2019;20:137–55.
Sim J, Johnson RS. Through a clear cell, darkly: HIF2alpha/PLIN2-maintained fat droplets protect ccRCCs from ER stress. Cancer Disco. 2015;5:584–5.
Welte MA. Expanding roles for lipid droplets. Curr Biol. 2015;25:R470–81.
Li Z, Johnson MR, Ke Z, Chen L, Welte MA. Drosophila lipid droplets buffer the H2Av supply to protect early embryonic development. Curr Biol. 2014;24:1485–91.
Bradley J, Pope I, Masia F, Sanusi R, Langbein W, Swann K, et al. Quantitative imaging of lipids in live mouse oocytes and early embryos using CARS microscopy. Development. 2016;143:2238–47.
Wang L, Zhang T, Wang L, Cai Y, Zhong X, He X, et al. Fatty acid synthesis is critical for stem cell pluripotency via promoting mitochondrial fission. EMBO J. 2017;36:1330–47.
Wu Y, Chen K, Liu X, Huang L, Zhao D, Li L, et al. Srebp-1 interacts with c-Myc to enhance somatic cell reprogramming. Stem Cells. 2016;34:83–92.
Bickel PE, Tansey JT, Welte MA. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta. 2009;1791:419–40.
Itabe H, Yamaguchi T, Nimura S, Sasabe N. Perilipins: a diversity of intracellular lipid droplet proteins. Lipids Health Dis. 2017;16:83.
Sztalryd C, Brasaemle DL. The perilipin family of lipid droplet proteins: gatekeepers of intracellular lipolysis. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862:1221–32.
Brasaemle DL, Rubin B, Harten IA, Gruia-Gray J, Kimmel AR, Londos C. Perilipin A increases triacylglycerol storage by decreasing the rate of triacylglycerol hydrolysis. J Biol Chem. 2000;275:38486–93.
Chiang HL, Terlecky SR, Plant CP, Dice JF. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246:382–5.
Xu Y, Zhang Y, Garcia-Canaveras JC, Guo L, Kan M, Yu S, et al. Chaperone-mediated autophagy regulates the pluripotency of embryonic stem cells. Science. 2020;369:397–403.
Kaushik S, Cuervo AM. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol. 2015;17:759–70.
Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–3.
Giacomello M, Pyakurel A, Glytsou C, Scorrano L. The cell biology of mitochondrial membrane dynamics. Nat Rev Mol Cell Biol. 2020;21:204–24.
Cogliati S, Enriquez JA, Scorrano L. Mitochondrial cristae: where beauty meets functionality. Trends Biochem Sci. 2016;41:261–73.
Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F, Kroemer G. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 2015;21:805–21.
Stein LR, Imai S. The dynamic regulation of NAD metabolism in mitochondria. Trends Endocrinol Metab. 2012;23:420–8.
He R, Dantas A, Riabowol K. Histone acetyltransferases and stem cell identity. Cancers. 2021;13:2407.
Kretsovali A, Hadjimichael C, Charmpilas N. Histone deacetylase inhibitors in cell pluripotency, differentiation, and reprogramming. Stem Cells Int. 2012;2012:184154.
Dancy BM, Cole PA. Protein lysine acetylation by p300/CBP. Chem Rev. 2015;115:2419–52.
Tasseva G, Bai HD, Davidescu M, Haromy A, Michelakis E, Vance JE. Phosphatidylethanolamine deficiency in Mammalian mitochondria impairs oxidative phosphorylation and alters mitochondrial morphology. J Biol Chem. 2013;288:4158–73.
Zhong X, Cui P, Cai Y, Wang L, He X, Long P, et al. Mitochondrial dynamics is critical for the full pluripotency and embryonic developmental potential of pluripotent stem cells. Cell Metab. 2019;29:979–92.e4.
Heden TD, Johnson JM, Ferrara PJ, Eshima H, Verkerke ARP, Wentzler EJ, et al. Mitochondrial PE potentiates respiratory enzymes to amplify skeletal muscle aerobic capacity. Sci Adv. 2019;5:eaax8352.
Arena R, Bisogno S, Gasior L, Rudnicka J, Bernhardt L, Haaf T, et al. Lipid droplets in mammalian eggs are utilized during embryonic diapause. Proc Natl Acad Sci USA. 2021;118:e2018362118.
Kaushik S, Cuervo AM. AMPK-dependent phosphorylation of lipid droplet protein PLIN2 triggers its degradation by CMA. Autophagy. 2016;12:432–8.
Khoa LTP, Tsan YC, Mao F, Kremer DM, Sajjakulnukit P, Zhang L, et al. Histone acetyltransferase MOF blocks acquisition of quiescence in ground-state ESCs through activating fatty acid oxidation. Cell Stem Cell. 2020;27:441–58.e10.
Lees JG, Gardner DK, Harvey AJ. Nicotinamide adenine dinucleotide induces a bivalent metabolism and maintains pluripotency in human embryonic stem cells. Stem Cells. 2020;38:624–38.
Hussein AM, Wang Y, Mathieu J, Margaretha L, Song C, Jones DC, et al. Metabolic control over mTOR-dependent diapause-like state. Dev Cell. 2020;52:236–50.e7.
Kratky D, Obrowsky S, Kolb D, Radovic B. Pleiotropic regulation of mitochondrial function by adipose triglyceride lipase-mediated lipolysis. Biochimie. 2014;96:106–12.
Matias MI, Yong CS, Foroushani A, Goldsmith C, Mongellaz C, Sezgin E, et al. Regulatory T cell differentiation is controlled by alphaKG-induced alterations in mitochondrial metabolism and lipid homeostasis. Cell Rep. 2021;37:109911.
Flis VV, Daum G. Lipid transport between the endoplasmic reticulum and mitochondria. Cold Spring Harb Perspect Biol. 2013;5:a013235.
Xuan Q, Hu C, Yu D, Wang L, Zhou Y, Zhao X, et al. Development of a high coverage pseudotargeted lipidomics method based on ultra-high performance liquid chromatography-mass spectrometry. Anal Chem. 2018;90:7608–16.
Acknowledgements
We thank all members in the lab of Dr Xingguo Liu for helpful suggestions. We also thank Shanghai ProfLeader Biotech Co., Ltd. for the assistance with isotope analysis.
Funding
This work was financially supported by the National Key Research and Development Program of China (2018YFA0107100), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16030505), the National Natural Science Foundation projects of China (92157202, 32025010, 98154001, 31900614, 31970709, 81901275, 32070729, 32100619, 32170747), the National Key Research and Development Program of China (2019YFA0904500, 2017YFA0106300, 2017YFA0102900), the Key Research Program, CAS (ZDBS-ZRKJZ-TLC003, QYZDB-SSW-SMC001), and International Cooperation Program, CAS (154144KYSB20200006), Guangdong Province Science and Technology Program (2020B1212060052, 2018GZR110103002, 2020A1515011200, 2020A1515010919, 2020A1515011410, 2021A1515012513, 2021B1515020096, 2022A1515012616), Guangzhou Science and Technology Program (202002030277, 202102020827, 202102080066), and CAS Youth Innovation Promotion Association (to YW and KC).
Author information
Authors and Affiliations
Contributions
Conceptualization: XL, YW, and KC; methodology: XL, YW, KC, and LL; investigation: YW, KC, LL, ZH, YL, GX, HY, HL, ZL, JL, CZ, JZ, DZ, and JW; formal analysis: YW, KC, LL, TW, JN, DY, GP, and WC; writing—original draft: XL and YW; writing—review and editing: XL, YW, and KC; funding acquisition: XL, YW, and KC; supervision: XL.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All the cells were obtained with approval from the ethics committee of the Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences (GIBH). All the animals were handled according to approved Institutional Animal Care and Use Committee protocols of GIBH.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by L. Scorrano
Rights and permissions
About this article
Cite this article
Wu, Y., Chen, K., Li, L. et al. Plin2-mediated lipid droplet mobilization accelerates exit from pluripotency by lipidomic remodeling and histone acetylation. Cell Death Differ 29, 2316–2331 (2022). https://doi.org/10.1038/s41418-022-01018-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-022-01018-8
This article is cited by
-
Aged mesenchymal stem cells and inflammation: from pathology to potential therapeutic strategies
Biology Direct (2023)
-
Bile acids-mediated intracellular cholesterol transport promotes intestinal cholesterol absorption and NPC1L1 recycling
Nature Communications (2023)
-
Harnessing metabolism of hepatic macrophages to aid liver regeneration
Cell Death & Disease (2023)
-
WD repeat domain 82 (Wdr82) facilitates mouse iPSCs generation by interfering mitochondrial oxidative phosphorylation and glycolysis
Cellular and Molecular Life Sciences (2023)
-
Functional differentiation of the ovine preadipocytes —insights from gene expression profiling
Functional & Integrative Genomics (2023)