Abstract
Tripartite motif-containing 28 (TRIM28) is an E3 ubiquitin ligase harboring multiple cellular functions. We found that the TRIM28 protein is frequently overexpressed in patients with lung cancer. The stable overexpression of TRIM28 in lung cancer cells and xenograft models significantly increased the proliferation, migration, and invasiveness, whereas knockdown of TRIM28 had the opposite effect. We further observed that TRIM28 regulates the ubiquitin ligases RLIM and MDM2 to target the p53 levels during lung tumorigenesis. These data provide new insights into lung cancer development and potential new therapeutic targets for this disease.
Similar content being viewed by others
Introduction
Lung cancer is one of the leading causes of cancer mortalities globally, resulting in more than one million deaths worldwide [1]. The 5-year survival rate in lung cancer is only approximately 15%, mainly due to the high late-stage diagnosis rates and metastatic potential [2]. Anticancer drugs such as gefitinib and erlotinib that target receptor tyrosine kinases (RTKs) or epidermal growth factor receptors (EGFRs) have shown some efficacy in these patients, but most non-small cell lung carcinoma (NSCLC) cases show drug resistance and metastatic propensities [3,4,5]
The lungs are exposed to various environmental stresses such as fine particulate matter and pathogens [6]. To maintain tissue homeostasis, cellular protein levels need to be tightly regulated. This protein quality control is mediated by the ubiquitin-proteasome system (UPS) and lysosomal/autophagic protein degradation pathways [7, 8]. The UPS regulates a variety of signaling pathways and is one of the major mechanisms controlling protein homeostasis and developmental decisions [6]. In the UPS pathways, ubiquitin is transferred to its targets via the E1–E2–E3 cascade, wherein the E3 ubiquitin ligases are responsible for substrate selection and therefore control ubiquitin chain linkage specificities [9,10,11,12].
TRIM28 (also known as KAP1 or TIF1-β) is a nuclear corepressor with various intracellular regulatory functions, including the responses to DNA damage and maintenance of stem cell pluripotency [13, 14]. TRIM28 also harbors a conserved RING finger domain and forms a complex with various proteins to target them for ubiquitination/degradation. TRIM28 has been shown to cooperate with MDM2 to regulate p53 ubiquitination and degradation in osteosarcoma cells [15].
RLIM (also known as RNF12) is another E3 ubiquitin ligase that maintains the expression of stem cell-specific genes and regulates X-chromosome inactivation [16,17,18]. RLIM interacts with MDM2 and mediates MDM2 degradation through UPS, which is independent of the self-ubiquitination activity of MDM2 [19].
To identify novel human lung cancer biomarkers, we examined the protein level changes in the tumor/non-tumor paired tissues from patients with lung cancer and found that TRIM28 is highly expressed in lung cancer tissues. We identified that RLIM is a novel substrate for the E3 ubiquitin ligase activity of TRIM28.
Furthermore, we showed that TRIM28 functions as an oncogene in lung cancer that downregulates the p53 protein levels in the p53–MDM2 cascade by targeting RLIM. Moreover, we analyzed the survival rates of patients with lung cancer and found that the levels of TRIM28 and RLIM correlate with overall survival. Thus, elucidating the precise regulatory mechanisms underlying the TRIM28–RLIM–MDM2–p53 cascade will provide new insights into lung cancer development and potential therapeutic strategies for this disease.
Materials and methods
Antibodies and reagents
The following antibodies were purchased from various vendors: PARP (9542), cleaved-caspase3 (9661, Cell Signaling Technology); HA-tag conjugated HRP (2999, Cell Signaling Technology); p21 (2947, Cell Signaling Technology); c-MYC (sc-40, Santa Cruz Biotechnology); ubiquitin (sc-8017, Santa Cruz Biotechnology); TRIM28 (sc-33186, Santa Cruz Biotechnology); β-actin (sc-47778, Santa Cruz Biotechnology; p53 (05–224, Millipore); MDM2 (ab16895, Abcam); anti-MYC-tag conjugated HRP (ab62928, Abcam); HA (MMS-101P, Covance); BAX (610982, BD Bioscience); and RNF12 (YF-MA11494, AbFrontier). DuoLink in situ fluorescence reagents were purchased from Sigma-Aldrich. Small-interfering RNAs (siRNAs) for human TRIM28 and RLIM were commercially synthesized by Dharmacon and Genolution, respectively, and the nucleotide sequences for these molecules are presented in Supplementary Table S1.
Patients and lung tissue samples
Written informed consent was obtained from all the patients with lung cancer who participated in this study. All experimental protocols were approved by the Institutional Review Board of Asan Medical Center and the University of Ulsan College of Medicine (2014-0960, 2020-1117). Human lung tissues were obtained through the Asan Bio Resource Center (2014-20[89]).
Animal studies
All mice were bred at the animal facilities of the Asan Institute for Life Sciences, University of Ulsan College of Medicine. All animal procedures were approved by the Institutional Ethics Committee and Institutional Animal Care Committee of University of Ulsan College of Medicine (2016-02-168 and 2017-12-281). The animal studies were conducted in a double-blinded manner by separating animal breeding and tissue analysis. The mouse experiments were repeated twice, and the numbers of animals in the experimental groups are described in the figure legends.
Cell culture
All cell lines (H460, H1299, HCC827, H2009, H1975, H3255, HEK293T, A549, and A549-Luc) were obtained from the American Type Culture Collection (ATCC). The H460, H1299, HCC827, H2009, H1975, and H3255 cells were cultured and passaged in Roswell Park Memorial Institute (RPMI) 1640 supplemented with 10% fetal bovine serum (FBS). The 293T, A549, and A549-Luc cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% FBS (Hyclone) and maintained in a humidified atmosphere of 5% CO2 at 37 °C.
Quantitative RT-PCR (qPCR)
Total RNAs were isolated using a TRIzol extraction kit following the manufacturer’s instructions (Invitrogen). cDNAs were then synthesized using SuperScript II Reverse Transcriptase (Invitrogen) and subjected to qPCR using SYBR Green I Master Mix on a CFX Connect Real-time PCR system (Bio-Rad). The qPCR primers used are listed in Supplementary Table S2.
Stable isotope labeling by amino acids in cell culture (SILAC)
Primary cells from lung cancer tissues and adjacent normal tissues were extracted from patients with lung cancer as described previously [20]. Primary cells (108) were grown in lysine free DMEM supplemented with 10% dialyzed FBS (Gibco), 2 mM l-glutamine (Thermo Fisher), pen/strep (Gibco), and light (K0) lysine (50 µg/ml). The heavy medium was replaced with K8-lysine. Heavy and light cells were mixed equally by cell number and lysed in 8 ml of denaturing lysis buffer (50 mM Tris pH 8.0, 8 M Urea, 75 mM NaCl supplemented with protease inhibitors [Roche]). The sonicated samples were diluted to lower the urea concentration to 4 M and digested with trypsin as described previously. Peptides were separated on 100 µm × 20 cm C18 reverse phase column with a gradient of 6 to 27% acetonitrile in 0.125% formic acid over 180 min. The 20 most intense peaks from each full MS scan acquired using the Orbitrap XL (Thermo) were selected for MS/MS.
Mass spectrometry
For proteomic analysis, retroviral vectors were used to stably express TRIM28 harboring a HA fusion tag in HeLa and A549 cells. These cells were then used in immunoprecipitation experiments, and any complexes formed were analyzed by mass spectrometry, as previously described [20]. Briefly, ~107 cells were treated with 125 mM DTBP (Pierce) for 10 min and neutralized for 5 min before being lysed in 4 ml of lysis buffer supplemented with protease inhibitors. Cleared lysates were filtered through 0.45-μm spin filters (Millipore) and immunoprecipitated with 30 μl of anti-HA resin (Sigma). The complexes were then washed with lysis buffer, resuspended in PBS, eluted with HA peptide, and precipitated with 10% trichloroacetic acid. The complexes were finally analyzed on a Thermo Orbitrap-XL mass spectrometer.
TCGA analysis
To compare the TRIM28 expression levels between normal and lung cancer tissues, RNAseq2 level 3 gene expression data provided by the Cancer Genome Atlas (TCGA) were analyzed. Normal data were obtained from 21 tumor-adjacent normal tissues including lung cancer-matched normal tissues. The expression values for TRIM28 were transformed to a log2 fold-change at a level normalized using RSEM [21].
Colony formation assay
Cells were seeded at a density of 2 × 103 cells per well in 6-well plates and cultured in DMEM for 2–3 weeks. The cells were then fixed with 3.7% paraformaldehyde/sucrose and stained with 0.5% crystal violet for 30 min.
Cell viability assay
Control or treated cells were seeded at a density of 1 × 105 cells per well and analyzed on days 1–4. At the end of each incubation period, CellTiter-Glo reagent (Promega) was added, and the plates were incubated at room temperature for 30 min on an orbital shaker. Luminescence intensities were determined using a GloMax® 96 Microplate Luminometer (Promega).
Wound-healing assay
Cells were seeded at a density of 5 × 105 cells per well in 6-well plates and incubated. Cell monolayers were mechanically wounded with a sterile 200-μl pipette tip and washed with medium to remove detached cells. Cell migration was evaluated by measuring the differences in the areas of the wounds.
Invasion assay
Cells were seeded at a density of 1 × 105 cells per well in the upper chamber of a Transwell insert (Corning) coated with 1–2 mg of Matrigel (BD Biosciences) that was placed in a 24-well plate. The lower chamber was filled with 800 μl of DMEM supplemented with 10% FBS, and the cells were incubated for 36 h. Cells that invaded through the Matrigel to the underside of the membrane were fixed with 3.7% paraformaldehyde/sucrose and stained with 0.5% crystal violet for 20 min. Cells that remained at the upper surface of the insert were removed with a cotton swab, and the cells that migrated through the upper chamber were counted in five random fields under a light microscope.
Generation of stable cell lines using plasmid and short hairpin RNA transfections
Lipofectamine 2000 (Invitrogen) or iN-fect (iNtRON) was used for the plasmid transfection of A549 and HEK293T cells following the manufacturer’s instructions. After 48 h, transfected cells were harvested for further analysis. For the generation of stable knockdown cell lines, miR-30–based short hairpin RNA (shRNA) constructs and VSVG and gag-pol helper vectors were used. Cells were selected for at least 1 week in puromycin or blasticidin. The shRNA sequences are listed in Supplementary Table S3.
Immunoblotting and immunoprecipitation
Human tissues were homogenized using a Dounce homogenizer in HEPES-buffered saline containing 10% glycerol, 1 mM EDTA, 2 mM EGTA, 1 mM DTT, 5 mM Na3VO4, and 5 mM NaF, supplemented with protease inhibitors (Roche). The cultured cells were rinsed once with ice-cold PBS and then lysed in lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 0.5% Triton X-100, and 1 mM EDTA) supplemented with protease inhibitors (Roche). Both sets of lysates were then centrifuged at 15000 g for 15 min, and the protein concentrations were determined by BCA assay (Pierce). Approximately 30 μg of protein extracts were resolved by SDS-PAGE, analyzed by western blot using primary antibodies followed by the corresponding IgG horseradish peroxidase-conjugated secondary antibodies (Thermo Fisher Scientific), and detected by enhanced chemiluminescence (Bio-Rad).
For the immunoprecipitation experiments, the clarified cell lysates (0.5–1 mg protein) were pre-cleared with 30 µl of protein-A/G sepharose 4 Fast Flow beads (GE Healthcare) for 1 h. The supernatant was incubated overnight with 3 µg of the appropriate antibody with rotation and then precipitated by mixing with 30 µl of protein-A/G beads at 4 °C for an additional 3 h. Anti-HA or anti-Myc antibody-conjugated agarose beads were used for immunoprecipitation of HA or Myc tagged proteins, respectively. The beads were washed five times with 1 ml of lysis buffer and then subjected to immunoblotting.
Construction of reporter plasmids and luciferase reporter assay
The PG13-Luc, a p53 reporter with a firefly luciferase gene under the control of 13 p53 response elements, was purchased from Addgene. A cDNA encoding Renilla luciferase was cloned into the pRL-SV40 vector (Promega). A549 and H1299 cells were transiently transfected with PG13-Luc and pRL-SV40 using Fugene HD transfection reagent (Roche). Cells were harvested and assayed for luciferase activities following the manufacturer’s instructions (Promega).
Plasmid construction and site-directed mutagenesis
All open reading frames (ORF) were cloned into CMV-MYC or CMV-HA vectors for transient transfection using Gateway technology (Invitrogen). For the domain-mapping experiment, the various TRIM28 and RLIM fragments were PCR-amplified and subcloned into vectors using the forward and reverse primer sets listed in Supplementary Table S4. All constructs and mutations were verified by nucleotide sequencing (IDT and Macrogen).
In vitro ubiquitylation assays
RLIM and TRIM28 proteins were purified from 293T cells using immobilized anti-HA or anti-Myc resin (Sigma), eluted with HA or Myc peptides (Sigma), and dialyzed against buffer (0.1% Nonidet P40, 1 mM DTT, 150 mM NaCl, 50 mM Tris-HCl pH 7.5, and 1 mM EDTA). RLIM ubiquitylation reactions were performed in reaction buffer containing 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 2 mM ATP, and 1 mM DTT. The indicated proteins were assembled on ice, and reaction was initiated by the addition of RLIM before incubation at 37 °C for 60 min. The reaction mixture contained 156 μM ubiquitin, 150 nM UBA1, 300 nM UBCH5, 350 nM TRIM28 (wild-type or mutant), and 4 μM RLIM-HA. At the end of the incubation, an equal volume of denaturing sample buffer was added, and the reactions were stopped by incubation at 95 °C for 7 min. The reaction products were examined by SDS-PAGE and immunoblotting with α-HA (RLIM) or α-MYC (TRIM28) antibodies.
Xenograft study
Male BALB/c nude mice (4–6 weeks old) were randomly divided into experimental groups. Mice were subcutaneously injected with A549 cells (1 × 106 cells), H1299 cells (2 × 106 cells), or A549-Luc-C8 cells (2.5 × 105 cells), as indicated in the figures, with a total volume of 50 ml using a 27-gauge needle. To determine the tumor mass in each animal, the greatest longitudinal diameter (length) and greatest transverse diameter (width) were measured using an external caliper. The tumor mass was then calculated by the modified ellipsoidal formula: V = (L × W2)/2, where V = volume (in mm3), L = length (in mm), and W = width (in mm). Bioluminescent imaging was carried out with IVIS Lumina Series III (Perkin Elmer). For each imaging session, the luciferin in PBS (a total of 150 mg luciferin per kg body weight) was administered into the peritoneal cavity according to the manufacturer’s protocol. Up to four animals were maintained in the instrument using the integral anesthetic manifold and imaged 10 min after the luciferin injection. The IVIS imaging system acquires a photographic image of the mice and a quantitative bioluminescent signal, which are then overlaid on the image.
Histology, immunohistochemistry, and immunofluorescence staining
Mice were anesthetized using isoflurane (N2O:O2/70%:30%) before transcardiac perfusion-fixation with heparinized saline containing 3.7% formaldehyde. The xenograft tumors were immediately embedded in OCT medium and frozen on dry ice. A cryotome was used to cut the tissue into 10-μm thick sections, which were placed on Superfrost Plus slides (Surgipath Medical), dried at room temperature, and kept at −80 °C until they were thawed for immunostaining.
For immunohistochemistry, paraffin-embedded sections of human lung tumor samples were de-waxed in xylene and rehydrated in ethanol. Antigen retrieval was subsequently performed by boiling the sections in a sodium citrate buffer (pH 6.0). Tissue sections were then incubated with indicated antibodies at 4 °C for 18 h. After washing three times with PBS, the sections were incubated with a peroxidase-conjugated secondary antibody and stained with 3′,3′-diaminobenzidine (DAB) substrate solution. The nuclei were further stained with hematoxylin. DAB-stained images were then obtained using the Leica DM 750 microscope.
For the TUNEL assay, frozen sections of xenograft tumors were incubated in PBS containing 0.1% Triton X-100. TUNEL reactions were then performed at 37 °C for 60 min using an In situ Cell Death Detection Kit, Fluorescein (Roche), following the manufacturer’s instructions. Cell nuclei were counterstained with DAPI. The tumor sections were mounted using Vectashield mounting medium, and fluorescence images were obtained in three random fields per tissue section at a ×100 magnification under an LSM 710 confocal microscope equipped with argon and helium-neon lasers (Zeiss).
Statistical analysis
All data were statistically analyzed using SPSS 16.0 (SPSS, Inc.), and the results are expressed as the mean ± standard deviation (SD) of at least three independent experiments. Comparisons between two groups were performed using Student’s t tests, whereas one- or two-way analysis of variance (ANOVA) with Dunnett’s post-hoc tests was used to compare the results of multiple groups. A P-value of <0.05 was considered significant.
Results
TRIM28 is highly expressed in lung cancer tissues
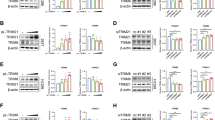
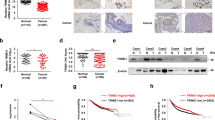
To identify new lung cancer markers, we analyzed the protein profiles between cancer and adjacent normal tissues from patients with lung cancer using stable isotope labeling with amino acids in cell culture-based quantitative proteomics (Fig. 1a). TRIM28 showed a significant ratio between cancer and normal tissues (Supplementary Fig. S1). To study the expression of TRIM28 in lung cancer, we analyzed the protein levels of TRIM28 in 104 paired lung cancer and adjacent normal tissues by western blotting (Fig. 1b, c and Supplementary Fig. S1) and conducted an immunohistochemical analysis using antibodies against TRIM28 and Ki67 (Fig. 1d). Notably, we found that TRIM28 was highly expressed in 79.8% of the samples (Fig. 1e) and that its expression levels were significantly increased in the lung cancer tissues (P < 0.001; Fig. 1e). Notably, TRIM28 proteins were overexpressed regardless of the type or stage of disease (Fig. 1f, Supplementary Table S4). Furthermore, consistent with our findings, the TCGA database indicated that the TRIM28 level is higher in lung cancer tissues than in normal tissues (Fig. 1g).
a Schematic overview of the SILAC quantitative proteomics approach used with paired lung cancer tissues and their adjacent normal lung tissues. b Representative immunoblot for TRIM28 and β-actin expression in 104 paired lung cancer tissues and their adjacent normal lung tissues. N, normal tissue; T, tumor. c Quantification of TRIM28 protein expression using immunoblotting analysis in 104 paired lung cancer tissues and their adjacent normal lung tissues. β-actin protein was used as an internal control. The error bar displays the SD value determined with a two-sided Student’s t test; ***P < 0.001. d Immunohistochemical analysis of tumor and normal lung tissues using antibodies against TRIM28 and Ki67. Scale bar, 50 μm. e Graph of TRIM28 protein expression determined by immunoblot analysis in 104 paired lung cancer tissues. β-actin protein was used as an internal control. f TRIM28 expression levels and clinical characteristics in the 104 lung cancer study cases. The P values represent probabilities for TRIM28 expression levels between variable subgroups determined using a two-sided Fischer’s exact-test (tumor node metastasis). g Expression levels of the TRIM28 gene in healthy individuals (normal, n = 640) and patients with lung cancer (tumor, n = 495) from the TCGA LUSC database. The box plot represents the average expression of TRIM28 (mean ± SD). The horizontal line indicates the median value.
Knockdown of TRIM28 inhibits tumor growth
To assess the effects of TRIM28 on lung cancer cell proliferation, multiple shTRIM28s were stably transfected into A549 cells (Fig. 2a). A reduction in TRIM28 expression level was associated with a significant decrease in cell proliferation (P < 0.001, t test; Fig. 2b, c). Additionally, a wound-healing assay in the TRIM28-knockdown cells showed significantly decreased cell migration (P < 0.001, t-test; Fig. 2d). Cell invasion assay showed that the knockdown of TRIM28 substantially suppressed the invasiveness of lung cancer cells (P < 0.001, t test; Fig. 2e). Similar results were obtained in H460 cells but not in H1299 cells (Supplementary Fig. S2). This may indicate that p53 is involved in the TRIM28 pathway in lung cancer because A549 and H460 lung adenocarcinoma cells harbor wild-type p53, whereas H1299 is a p53-null lung cancer cell line [22].
a Immunoblotting analysis of TRIM28 in A549 cells at 48 h after its knockdown using four lentiviral shRNAs. b Effects of the TRIM28 knockdown on the colony formation of A549 cells. A549 cells transfected with TRIM28 shRNAs were cultured for 10 days and stained with crystal violet to visualize and count the colonies. c Effects of the TRIM28 knockdown on the viability of A549 human lung cancer cells. Cell viability was monitored for 3 days using a CellTiter-Glo luminescent assay (normalized to control wild-type cells at day 0; **P < 0.01). d Effects of the TRIM28 knockdown on the migration of A549 lung cancer cells in a wound-healing assay. Wound closure was determined in control and TRIM28-knockdown cells at 24-, 48-, and 72-h time points. (***P < 0.001). e Effects of the TRIM28 knockdown on cell invasion. A549 cells with stably knocked down TRIM28 and controls were assayed in Transwell inserts containing Matrigel-coated membranes. Invasive potential was assessed after a 24-h incubation. Values shown are the mean ± SD of the number of invading cells (***P < 0.001). Scale bar, 100 μm. f A549 cells with stably knocked down TRIM28 and controls were subcutaneously injected into athymic (nu/nu) mice and tumor growth was measured using the formula 0.52 × length × width2 every 3 days after injection. Tumor sizes are indicated by the volume, and the graph shows the average volume ± SD of implanted tumors (n = 7 or 8 mice per group, ***P < 0.001). All data values are expressed as the mean ± SD of the results obtained from three independent experiments. A two-sided Student’s t test was used for statistical analysis.
Next, to assess the effects of TRIM28 on tumor growth in vivo, A549 or H1299 cells stably expressing control or shTRIM28s were subcutaneously injected into the lower left flank of NOD-SCID mice. Consistent with our in vitro results, TRIM28-knockdown mice had a significantly lower tumor volume than the control mice (mean tumor volume on day 30 for control vs. TRIM28, 338.74 mm3, SD = 38.67 mm3 vs. 41.39 mm3, SD = 24.17 mm3; P < 0.001; Fig. 2f); this was observed in those transfected with A549 cells but not H1299 cells (Supplementary Fig. 3). These data further supported the critical role of TRIM28 in lung cancer cell growth and the involvement of p53 in this pathway.
TRIM28 overexpression promotes tumorigenic growth in the lung
To further verify that TRIM28 is involved in lung cancer cell proliferation, migration, and invasion, we stably overexpressed TRIM28 in A549 cells and performed similar assays to those used in the knockdown experiments (Fig. 3a–f). In contrast to the TRIM28-knockdown cells, the results of colony formation, cell proliferation, and migration and invasion assays revealed a marked elevation of these properties in TRIM28-overexpressed cells (P < 0.01, P < 0.01, P < 0.01, and P < 0.001, t test, respectively; Fig. 3b–e). To assess the effects of TRIM28 on tumor formation in vivo, stable TRIM28-expressing A449 cells were subcutaneously injected into NOD-SCID mice. As shown in Fig. 3f, the TRIM28-overexpression mice had statistically larger tumor volumes than the control mice (mean tumor volume on day 24, control vs. TRIM28, 293.12 mm3, SD = 37.77 mm3, vs. 618.84 mm3, SD = 59.63 mm3; P < 0.001; Fig. 3f). Taken together, the overexpression and knockdown results provide further support for the critical role of TRIM28 in lung tumor growth.
a Immunoblotting analysis of A549 cells stably expressing TRIM28. b Effects of TRIM28 overexpression on colony formation. A549 cells overexpressing TRIM28 were grown for 10 days and the colonies were stained with crystal violet and counted. Data shown in the graph represent the average colony number ± SD (**P < 0.01). c Effects of TRIM28 overexpression on the viability of A549 human lung cancer cells which was monitored for 4 days using a CellTiter-Glo luminescent assay (with normalization to control wild-type cells at day 0, **P < 0.01). d Effects of TRIM28 overexpression on the migration of A549 lung cancer cells in a wound-healing assay. Wound closure was determined in control and TRIM28 overexpressing cells at 12-, 24-, and 48-h time points. (**P < 0.01). e Effects of TRIM28 overexpression on cell invasion. Vehicle or TRIM28-overexpressing A549 cells were assayed in Transwell inserts containing a Matrigel-coated membrane. Invasive potential was assessed after a 24-h incubation. Values are expressed as the mean ± SD of the number of invading cells (**P < 0.01). Scale bar, 100 μm. f A549 cells stably expressing control or exogenous TRIM28-expressing vector were subcutaneously injected into athymic (nu/nu) mice and tumor growth was measured using the formula 0.52 × length × width2 every 3 days after injection. Tumor sizes are indicated by the volume, and the graph shows the average volume ± SD of implanted tumors (n = 7 or 8 mice per group, ***P < 0.01). All data values are expressed as the mean ± SD of the results obtained from three independent experiments. A two-sided Student’s t test was used for statistical analysis.
RLIM acts as a TRIM28-interacting protein
To further dissect the mechanisms involving TRIM28 in lung cancer development, we identified putative substrates for this factor using interaction proteomics (Fig. 4a). RLIM was identified as a high-confidence candidate protein interacting with most other peptides (Supplementary Table S2). To validate this putative physical association between TRIM28 and RLIM, we analyzed the expression of endogenous proteins in A549 and H1299 cells. Both proteins were detectable in anti-TRIM28 immunoprecipitated complexes but not in the control (Fig. 4b). Next, we examined the transient co-expression of HA-TRIM28 and Myc-RLIM in A549, H1299, and 293T cells. Immunoprecipitation of HA-TRIM28 complexes from the total cell lysates confirmed the interaction between TRIM28 and RLIM (Fig. 4c and Supplementary Fig. S4). Moreover, a proximity ligation assay also confirmed the TRIM28-RLIM interaction in the nucleus of A549 cells (Fig. 4d).
a Flow chart of the mass spectrometric analysis of A549 cells stably expressing HA-TRIM28. b Extracts from A549 and H1299 cells were immunoprecipitated with either control IgG or TRIM28 antibodies, and then the presence of RLIM and TRIM28 was determined by immunoblotting. c Extracts from A549 and H1299 cells expressing HA-TRIM28 and either empty vector or Myc-RLIM were immunoprecipitated with HA-conjugated antibodies, and IP complexes, and total cell lysates (TCL) were then immunoblotted with the indicated antibodies. d Cellular localization of TRIM28 and RLIM. A proximal ligation assay (in situ PLA) to detect binding between TRIM28 and RLIM proteins was performed in A549 cells using DuoLink® in situ probes with the indicated antibodies. The binding of anti-TRIM28 and anti-RLIM antibodies was detected by emission of red fluorescence. e Schematic overview of TRIM28 domains and deletion mutants. f Schematic overview of RLIM domains and deletion mutants. g Bacterially expressed HA-TRIM28 and various deletion mutants were bound to glutathione-sepharose beads and incubated with Myc-RLIM proteins. Bound Myc-RLIM proteins were detected by immunoblotting with anti-Myc antibody. h Bacterially expressed Myc-RLIM and various deletion mutants were bound to glutathione-sepharose beads and incubated with HA-TRIM28 proteins. Bound HA-TRIM28 proteins were detected by immunoblotting with anti-HA antibody.
To map the domain that interacts with RLIM, we generated a series of truncation mutants of TRIM28 (Fig. 4e). TRIM28 protein consists of four major structural domains—RBCC domain (comprising the RING [Really Interesting New Gene] domain, two B-box type domains, and a leucine zipper coiled-coil domain), HP1 protein binding domain, Plant homeodomain, and Bromodomain [13]. A pull-down assay with HA-TRIM28 and its various truncation mutants was used to assess their interactions with Myc-RLIM. Western blotting revealed that one discrete N-terminal domain of TRIM28 containing RBCC was critical for RLIM binding (Fig. 4g). We also performed a co-immunoprecipitation assay to examine the binding of TRIM28 to RLIM variants. A deletion mutant of RLIM containing amino acids 1 to 312 showed a strong association with HA-TRIM28 (Fig. 4h).
TRIM28 regulates RLIM through the ubiquitin-proteasome system
Since both TRIM28 and RLIM are E3 ubiquitin ligases and interact with each other, we hypothesized that TRIM28 regulates the ubiquitin-dependent degradation of RLIM or vice versa. We first observed that TRIM28 knockdown significantly increased the RLIM protein levels in various cell lines (Fig. 5a and Supplementary Fig. S5a). In contrast, RLIM depletion did not alter the TRIM28 protein levels (Fig. 5b). Next, we examined the turnover of RLIM in the absence of TRIM28. Knockdown of TRIM28 significantly increased the half-life of RLIM in A549 and H1299 cells, indicating that TRIM28 maintains the turnover of RLIM (Fig. 5c, d). We also tested whether TRIM28 promotes RLIM ubiquitylation in A549 cells. Treatment with shTRIM28s stabilized the RLIM protein and significantly decreased the length of its polyubiquitin chain in A549 cells (Fig. 5e). Additionally, we found that TRIM28, but not the mutant form of TRIM28, promotes RLIM ubiquitination in vitro, which is consistent with findings in mammalian cells (Fig. 5f).
a RLIM protein levels in A549, H460, and H1299 cells transfected with TRIM28-1 and -2 shRNA constructs. Cells were treated with siRNAs and the samples were immunoblotted with RLIM, TRIM28, and β-actin antibodies. b TRIM28 protein levels in A549, H460, and H1299 cells transfected with RLIM-1 and -2 shRNA constructs. Cells were treated with siRNAs and immunoblotted with RLIM, TRIM28, and β-actin antibodies. c RLIM protein stability assessed by immunoblotting. A549 and H1299 cells were transfected for 72 h with the TRIM28-1 and -2 shRNA constructs followed by incubation with 50 μg/ml cycloheximide (CHX) for the indicated times. Cell lysates were prepared and immunoblotted with anti-RLIM, TRIM28, and β-actin antibodies. d Graphs indicating the relative amounts of RLIM protein compared with the levels of actin in the immunoblot. The expression of each protein was analyzed using ImageJ. A two-sided Student’s t test was used for the statistical analysis; **P < 0.01, ***P < 0.001. e A549 cells were transfected with TRIM28 or control siRNAs. Some cells were treated with velcade for 5 h before harvesting. Cell lysates were prepared, and the RLIM-ubiquitin conjugates were detected by immunoblotting with an anti-RLIM antibody. f Increasing amounts of HA-TRIM28 (or its C65A variant with no enzymatic activity) was co-transfected with equivalent amounts of Myc-RLIM into A549 cells. After 48 h, cell lysates were prepared for immunoblotting. g A549 cells were transfected with mixtures of plasmids expressing HA-TRIM28 and MYC-RLIM followed by treatment with cycloheximide (CHX). Some cells were treated with velcade for 5 h before harvesting. The cell lysates were immunoblotted with anti-MYC, -HA, and -β-actin antibodies. f In vitro ubiquitination of RLIM. Purified ubiquitin, E1, E2, RLIM, and TRIM28 or TRIM28 (C65A) were incubated, and samples were resolved by SDS-PAGE and subjected to immunoblot analysis as indicated antibodies.
Next, we examined whether TRIM28 affects the RLIM protein level through its RING domain. A549 cells were co-transfected with vectors encoding Myc-RLIM and increasing amounts of the HA-TRIM28 or HA-TRIM28 C65A mutant devoid of catalytic activity. The protein levels of RLIM gradually decreased upon the co-expression of TRIM 28 in high amounts but not TRIM28 C65A (Supplementary Fig. S5b). In addition, the TRIM28 C65A mutant successfully interacted with RLIM protein but did not affect its levels in 293 T cells (Supplementary Fig. S5c). Moreover, TRIM28 C65A had no effect on RLIM half-life (Supplementary Fig. S5d). These results suggest that the N-terminal RING domain of TRIM28 is responsible for the protein stability of RLIM. Thus, our current data demonstrate that TRIM28 promotes RLIM ubiquitylation and degradation in a RING domain-dependent manner.
TRIM28 regulates the p53 protein levels through RLIM-MDM2-dependent degradation
A previous study reported that RLIM regulates p53 through MDM2 degradation [19]. Thus, we hypothesized that TRIM28 may regulate the p53 protein through the RLIM and MDM2 sequential ubiquitin ligase enzyme cascades. First, we examined the turnover of MDM2 and p53 in the absence of TRIM28, or TRIM28 and RLIM. When TRIM28 levels were depleted in A549 and H460 lung cancer cell lines, the half-life of RLIM was significantly increased as expected. However, the levels of MDM2 were drastically decreased, but those of p53 protein were increased. Conversely, the turnover rates of MDM2 and p53 were reversed when both TRIM28 and RLIM were knocked down in A549 and H460 cells (Fig. 6a–c). This indicates that TRIM28 regulates the p53 protein through RLIM and MDM2 degradation. Similar results were observed in H1975, H3255, HCC827, and H2009 cells (Supplementary Fig. S6a).
a Western blot analysis of A549 and H460 cells after siRNA-mediated TRIM28 knockdown. b A549 and H460 cells were treated with the indicated siRNAs followed by cycloheximide (CHX) for the indicated times. For assessment of the half-life of MDM2-p53 protein complexes, cell lysates were collected and immunoblotted with the indicated antibodies. c The graphs indicate the relative amounts of MDM2 and p53 proteins compared with the levels of actin in the blot. The expression of each protein signal was analyzed using ImageJ software. d–e A549 (p53 WT) and H1299 (p53 null) cells were transfected with PG3 promoter to assess the p53 activity under conditions of TRIM28 knockdown or TRIM28/RLIM double knockdown for 16 h. Cells were then lysed and assayed for luciferase activities. The values shown are the mean ± SD fold increases normalized using control siRNA cells (**P < 0.01, ***P < 0.001). f–g A549 cells were transfected with TRIM28, TRIM28/RLIM, or control siRNAs. The endogenous p21, Bax, p53, PARP, and cleaved caspase-3 levels were then determined by immunoblotting.
Next, we examined whether TRIM28 or TRIM28/RLIM knockdown affected p53 activity using a PG13 promoter assay. When shTRIM28s were transfected into p53 wild-type A549 cells, we observed a marked increase in the p53 level, but this effect was reversed by the knockdown of both TRIM28 and RLIM (Fig. 6d). Similar results were obtained when we introduced p53 into p53-null H1299 cells (Fig. 6e). To assess the effects of the TRIM28-RLIM axis on p53 downstream factors, TRIM28 and TRIM28/RLIM were knocked down in A549 cells. The TRIM28 knockdown increased the levels of p53 target proteins such as p21 and Bax, but this effect was reversed by a combined TRIM28/RLIM knockdown (Fig. 6f and Supplementary Fig. S7a). We also confirmed that increased p21 and Bax levels led to an increase in the expression of the apoptotic gene PARP and cleaved caspase-3 (Fig. 6g). Quantitative real-time PCR produced similar results for p53, p21, MDM2, PUMA, BAX, and NOXA upon TRIM28- or TRIM28/RLIM-knockdown but not in p53-null H1299 cells (Fig. S7b, c). In summary, our data confirmed that TRIM28 promotes MDM2-dependent p53 ubiquitination/degradation through RLIM.
Effects of a combined TRIM28/RLIM knockdown in a xenograft model and TRIM28-RLIM protein levels in patients with lung cancer
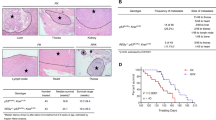
We confirmed that the TRIM28-RLIM axis regulates tumor activity in a xenograft model. Knockdown of TRIM28 significantly inhibited tumor growth in mice while double knockdown of TRIM28 and RLIM reversed this effect (Fig. 7a, b), which further indicated that TRIM28 regulates the RLIM protein during tumorigenesis. Analysis of apoptosis in tumor sections from patients with lung cancer showed consistent results with the xenograft experiments (Supplementary Fig. S8). We finally verified whether the TRIM28 and RLIM protein levels affected tumorigenesis in patients with lung cancer by analyzing their expression levels in 104 paired lung cancer and adjacent normal tissues by western blotting (Fig. 7c and Supplementary Fig. S9). Notably, the survival analysis revealed that patients with high TRIM28 and low RLIM levels had poorer overall survival than those with high TRIM28 and high/unchanged RLIM levels, which is consistent with our data (Fig. 7d). Collectively, the findings of our study indicate that the TRIM28/RLIM protein levels may be feasible prognostic markers for lung cancer and that these factors may be potential therapeutic targets for this disease.
a-b A549-luc-C8 cells stably expressing shTRIM28 or shRLIM were subcutaneously injected into the mice, and tumor sizes were measured at the indicated time points. Data in the graph show the quantification of the bioluminescent images. A two-sided Student’s t test was used for the statistical analysis (**P < 0.01). c Representative immunoblot of TRIM28-RLIM and β-actin expression in 104 paired lung cancer and adjacent normal lung tissues (N, normal; T, tumor). d Kaplan–Meier curve for the overall survival probability of the 101 lung cancer study patients corresponding to the TRIM28 and RLIM expression levels. P values were calculated using the log-rank test. e Proposed model.
Discussion
Our studies have shown that TRIM28 mediates lung tumorigenesis by targeting the E3 ubiquitin ligase RLIM (RNF12) in the p53-MDM2 axis. Furthermore, the E3 ligase activity of TRIM28 is involved in the degradation of RLIM, an MDM2 inhibitor, which maintains low p53 protein levels (Fig. 7e). This in turn promotes tumor cell proliferation and survival through the inhibition of p53 response elements such as p21, a CDK inhibitor [23], and Bax, an apoptotic protein [24].
To identify differentially expressed proteins during lung cancer development, we investigated the changes in protein levels in tumor-normal tissue pairs from 106 patients with lung cancer and found that TRIM28 is highly expressed in lung cancer tissues (Fig. 1). Consistent with our findings, several other groups have also reported that TRIM28 is highly expressed in various cancer types including lung cancer [25,26,27,28]. Overexpression of TRIM28 stimulated the tumorigenic activities such as proliferation, migration, and invasion in A549 cells (Fig. 3a–e). Additionally, when A549/TRIM28 cells were injected into nude mice, the subsequent xenograft experiments revealed that TRIM28 protein levels are critical for tumor development. To further confirm the tumorigenic role of TRIM28, we performed knockdown experiments using shRNA constructs with different target sequences in A549 cells (Fig. 2). Subsequent cell-based analyses such as colony formation and wound-healing assays revealed that a deficiency in TRIM28 suppresses the proliferation, migration, and invasiveness of the A549 cells (Fig. 2a–e). Furthermore, xenograft experiments with TRIM28 knockdown indicated that a reduction in the TRIM28 levels has a marked anti-tumorigenic effect (Fig. 2f).
To better understand the tumor growth mechanisms involving TRIM28 in lung cancer cells, we searched for TRIM28-binding proteins using co-immunoprecipitation and LC-MS/MS analyses. We thereby identified RLIM, which is an E3 ubiquitin ligase originally isolated as a LIM-homeodomain-binding protein [29]. We thus examined the possible relationship between TRIM28 and RLIM proteins in the p53-MDM2 axis. Results revealed that RLIM is a downstream target of TRIM28 and functions between TRIM28 and MDM2 and thereby acts as an MDM2 inhibitor to regulate p53 protein levels. The E3 ubiquitin ligase activity of TRIM28 is critical for mediating this TRIM28-dependent RLIM poly-ubiquitination and degradation (Figs. 5, 6).
In lung tumors with low p53 levels, RLIM may be induced by a deficiency in p53 levels to degrade MDM2 through a positive feedback mechanism. However, in the case of patients with lung cancer with high TRIM28 expression, TRIM28 could degrade RLIM in a cell-autonomous manner to maintain low p53 levels and thereby promote tumor cell proliferation and survival. Therefore, lung cancers with high TRIM28 levels can develop more severe malignancies than those with low TRIM28 levels.
Previous studies have shown that the nuclear co-repressor TRIM28 interacts with MDM2 and their interaction stimulates p53–HDAC1 complex formation, suppresses p53 acetylation, and facilitates p53 ubiquitination, subsequently inhibiting its transcriptional activity and regulatory functions [15, 30]. This is consistent with our results that the regulation of p53 degradation initiated by TRIM28 but it may be slightly different in lung cancer since RLIM plays a crucial role in p53 ubiquitination and degradation in vitro and in vivo (Fig. 6, Fig. 7a, b).
Hence, a better understanding of the extrinsic regulatory pathways of the TRIM28-RLIM-MDM2-p53 cascade is vital for elucidating the processes underlying lung cancer development. Indeed, RLIM acts as a component of the transforming growth factor β (TGF-β) signaling pathway [31]. The cytokine TGF-β induces the migration of lung cancer cells [32]. In addition, the development of therapeutic antibodies against TRIM28 or small molecules that inhibit the interaction of TRIM28 and RLIM may be viable new treatment approaches for patients with lung cancer with high TRIM28 expression.
Taken together, we demonstrated that TRIM28 modulates the p53-MDM2 axis in lung tumorigenesis by targeting RLIM through its E3 ubiquitin ligase activity. Revealing the precise regulatory mechanisms underlying this TRIM28-RLIM-MDM2-p53 cascade will provide new insights into lung cancer development and novel therapeutic approaches.
References
Didkowska J, Wojciechowska U, Manczuk M, Lobaszewski J. Lung cancer epidemiology: contemporary and future challenges worldwide. Ann Transl Med. 2016;4:150.
Fernandez HR, Gadre SM, Tan M, Graham GT, Mosaoa R, Ongkeko MS, et al. The mitochondrial citrate carrier, SLC25A1, drives stemness and therapy resistance in non-small cell lung cancer. Cell Death Differ. 2018;25:1239–58.
Rosell R, Karachaliou N, Arrieta O. Novel molecular targets for the treatment of lung cancer. Curr Opin Oncol. 2020;32:37–43.
Tan C-S, Gilligan D, Pacey S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol. 2015;16:e447–e59.
Minguet J, Smith KH, Bramlage P. Targeted therapies for treatment of non-small cell lung cancer – recent advances and future perspectives. Int J Cancer. 2016;138:2549–61.
Weathington NM, Mallampalli RK. New insights on the function of SCF ubiquitin E3 ligases in the lung. Cell Signal. 2013;25:1792–8.
Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol cell Biol. 2005;6:79–87.
Chen J, Chen ZJ. Regulation of NF-kappaB by ubiquitination. Curr Opin Immunol. 2013;25:4–12.
Wickliffe Katherine E, Lorenz S, Wemmer David E, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144:769–81.
Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–64.
Jin J, Li X, Gygi SP, Harper JW. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature 2007;447:1135–8.
Lee PC, Sowa ME, Gygi SP, Harper JW. Alternative ubiquitin activation/conjugation cascades interact with N-end rule ubiquitin ligases to control degradation of RGS proteins. Mol Cell. 2011;43:392–405.
Czerwinska P, Mazurek S, Wiznerowicz M. The complexity of TRIM28 contribution to cancer. J Biomed Sci. 2017;24:63.
Chen L, Munoz-Antonia T, Cress WD. Trim28 contributes to EMT via regulation of E-cadherin and N-cadherin in lung cancer cell lines. PloS ONE. 2014;9:e101040.
Wang C, Ivanov A, Chen L, Fredericks WJ, Seto E, Rauscher FJ 3rd, et al. MDM2 interaction with nuclear corepressor KAP1 contributes to p53 inactivation. EMBO J. 2005;24:3279–90.
Bustos F, Segarra-Fas A, Chaugule VK, Brandenburg L, Branigan E, Toth R, et al. RNF12 X-linked intellectual disability mutations disrupt E3 ligase activity and neural differentiation. Cell Rep. 2018;23:1599–611.
Gao R, Wang L, Cai H, Zhu J, Yu L. E3 ubiquitin ligase RLIM negatively regulates c-Myc transcriptional activity and restrains cell proliferation. PLoS ONE. 2016;11:e0164086.
Wang F, Bach I. Rlim/Rnf12, Rex1, and X chromosome inactivation. Front Cell Dev Biol. 2019;7:258.
Gao K, Wang C, Jin X, Xiao J, Zhang E, Yang X, et al. RNF12 promotes p53-dependent cell growth suppression and apoptosis by targeting MDM2 for destruction. Cancer Lett. 2016;375:133–41.
Kim SJ, Lee TH, Nam SH, Kim JH, Oh S, Cho YS, et al. Association of Uba6-Specific-E2 (USE1) With Lung Tumorigenesis. J Natl Cancer Inst. 2017;109:1–11.
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323.
Wang T, Li K, Song H, Xu D, Liao Y, Jing B, et al. p53 suppression is essential for oncogenic SPAG5 upregulation in lung adenocarcinoma. Biochem Biophys Res Commun. 2019;513:319–25.
Vilgelm AE, Saleh N, Shattuck-Brandt R, Riemenschneider K, Slesur L, Chen SC, et al. MDM2 antagonists overcome intrinsic resistance to CDK4/6 inhibition by inducing p21. Sci Transl Med. 2019;11:eaav7171.
Gajendran B, Durai P, Madhu Varier K, Chinnasamy A. A novel phytosterol isolated from Datura inoxia, RinoxiaB is a potential cure colon cancer agent by targeting BAX/Bcl2 pathway. Bioorgan. Med Chem. 2020;28:115242.
Liu L, Zhang L, Wang J, Zhao X, Xu Q, Lu Y, et al. Downregulation of TRIM28 inhibits growth and increases apoptosis of nude mice with nonsmall cell lung cancer xenografts. Mol Med Rep. 2018;17:835–42.
Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11:792–804.
Wang Y, Jiang J, Li Q, Ma H, Xu Z, Gao Y. KAP1 is overexpressed in hepatocellular carcinoma and its clinical significance. Int J Clin Oncol. 2016;21:927–33.
Liu L, Zhao E, Li C, Huang L, Xiao L, Cheng L, et al. TRIM28, a new molecular marker predicting metastasis and survival in early-stage non-small cell lung cancer. Cancer Epidemiol. 2013;37:71–8.
Zhang L, Huang H, Zhou F, Schimmel J, Pardo CG, Zhang T, et al. RNF12 controls embryonic stem cell fate and morphogenesis in zebrafish embryos by targeting Smad7 for degradation. Mol cell. 2012;46:650–61.
Okamoto K, Kitabayashi I, Taya Y. KAP1 dictates p53 response induced by chemotherapeutic agents via Mdm2 interaction. Biochem Biophys Res Commun. 2006;351:216–22.
Hill CS. Inhibiting the inhibitor: the role of RNF12 in TGF-beta superfamily signaling. Mol cell. 2012;46:558–9.
Hedrick E, Mohankumar K, Safe S. TGFbeta-induced lung cancer cell migration is NR4A1-dependent. Mol Cancer Res: MCR. 2018;16:1991–2002.
Acknowledgements
The authors thank the Optical Imaging and Optical Microscopy Cores at the Convergence Medicine Research Center (CREDIT), Asan Medical Center, for support and instruments.
Funding
This study was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MIST) (2019R1A2C2084181, 2020R1A4A1016029).
Author information
Authors and Affiliations
Contributions
J.J., G.L., S.N., T.L., D.K., and J.Y. conceived and designed analysis; J.J., G.L., and P.L. performed development of methodology and writing, review and revision of the paper. All authors read and approved the final paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
Written informed consent was obtained from all the patients with lung cancer who participated in this study. All experimental protocols were approved by the Institutional Review Board of Asan Medical Center and the University of Ulsan College of Medicine (2014-0960, 2020-1117). The study was performed in accordance with the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by M. Oren
Rights and permissions
About this article
Cite this article
Jin, JO., Lee, G.D., Nam, S.H. et al. Sequential ubiquitination of p53 by TRIM28, RLIM, and MDM2 in lung tumorigenesis. Cell Death Differ 28, 1790–1803 (2021). https://doi.org/10.1038/s41418-020-00701-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-020-00701-y
This article is cited by
-
DCAF13 inhibits the p53 signaling pathway by promoting p53 ubiquitination modification in lung adenocarcinoma
Journal of Experimental & Clinical Cancer Research (2024)
-
POLE2 promotes osteosarcoma progression by enhancing the stability of CD44
Cell Death Discovery (2024)
-
N6-methyladenosine hypomethylation of circGPATCH2L regulates DNA damage and apoptosis through TRIM28 in intervertebral disc degeneration
Cell Death & Differentiation (2023)
-
TRIM28 promotes the escape of gastric cancer cells from immune surveillance by increasing PD-L1 abundance
Signal Transduction and Targeted Therapy (2023)
-
Deer antlers: the fastest growing tissue with least cancer occurrence
Cell Death & Differentiation (2023)