Abstract
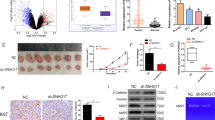
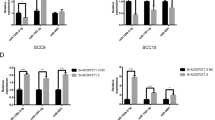
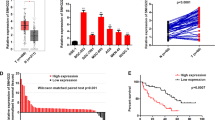
Increasing evidence proved the abnormal expression of long non-coding RNAs (lncRNAs) in various human malignancies, including oral squamous cell carcinoma (OSCC). Nevertheless, limited explorations concern the role of lncRNA small nucleolar RNA host gene 17 (SNHG17) in OSCC. Herein, SNHG17 was disclosed to be remarkably upregulated in OSCC cell lines and promoted OSCC cell growth. Further mechanistic studies, including DNA/RNA pull down, RIP, ChIP, and luciferase reporter gene assays, were conducted. It was confirmed that Wnt/β-catenin signaling pathway was involved in the SNHG17-mediated OSCC cell growth. Moreover, E74 like ETS transcription factor 1 (ELF1) was identified as the transcription activator of CTNNB1 (β-catenin mRNA) in OSCC. Inspired by competing for endogenous RNAs (ceRNAs) network, we were pleasantly surprised to find that SNHG17 and ELF1 functioned as ceRNAs in OSCC via competitively binding to microRNA-384 (miR-384). By using rescue assays, we revealed that SNHG17 facilitated OSCC cell growth through modulating miR-384/ELF1 axis. Importantly, we certified that ELF1 was indispensable for SNHG17-affected OSCC progression. Collectively, it can be concluded that SNHG17/miR-384/ELF1 axis contributed to OSCC cell growth via promoting CTNNB1 expression, thus activating Wnt/β-catenin signaling pathway.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Overgaard J, Hansen HS, Specht L, Overgaard M, Grau C, Andersen E, et al. Five compared with six fractions per week of conventional radiotherapy of squamous-cell carcinoma of head and neck: DAHANCA 6 and 7 randomised controlled trial. Lancet. 2003;362:933–40.
Zheng M, Jiang YP, Chen W, Li KD, Liu X, Gao SY, et al. Snail and Slug collaborate on EMT and tumor metastasis through miR-101-mediated EZH2 axis in oral tongue squamous cell carcinoma. Oncotarget. 2015;6:6797–810.
Kessler P, Grabenbauer G, Leher A, Bloch-Birkholz A, Vairaktaris E, Neukam FW. Neoadjuvant and adjuvant therapy in patients with oral squamous cell carcinoma long-term survival in a prospective, non-randomized study. Br J Oral Maxillofac Surg. 2008;46:1–5.
Pérez-Sayáns M, Suárez-Peñaranda JM, Pilar GD, Barros-Angueira F, Gándara-Rey JM, García-García A. Hypoxia-inducible factors in OSCC. Cancer Lett. 2011;313:1–8.
Costa FF. Non-coding RNAs: new players in eukaryotic biology. Gene. 2005;357:83–94.
Cui M, Zheng M, Sun B, Wang Y, Ye L, Zhang X. A long noncoding RNA perturbs the circadian rhythm of hepatoma cells to facilitate hepatocarcinogenesis. Neoplasia. 2015;17:79–88.
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6.
Zhang F, Ruan X, Ma J, Liu X, Zheng J, Liu Y, et al. DGCR8/ZFAT-AS1 promotes CDX2 transcription in a PRC2 complex-dependent manner to facilitate the malignant biological behavior of glioma cells. Mol Ther. 2020;28:613–30.
Li Z, Qin X, Bian W, Li Y, Shan B, Yao Z, et al. Exosomal lncRNA ZFAS1 regulates esophageal squamous cell carcinoma cell proliferation, invasion, migration and apoptosis via microRNA-124/STAT3 axis. J Exp Clin Cancer Res. 2019;38:477.
Jiang H, Li Y, Li J, Zhang X, Niu G, Chen S, et al. Long noncoding RNA LSINCT5 promotes endometrial carcinoma cell proliferation, cycle, and invasion by promoting the Wnt/β-catenin signaling pathway via HMGA2. Ther Adv Med Oncol. 2019;11:1758835919874649.
Semenza GL. The hypoxic tumor microenvironment: a driving force for breast cancer progression. Biochim Biophys Acta. 2016;1863:382–91.
Palmieri F, Monné M. Discoveries, metabolic roles and diseases of mitochondrial carriers: a review. Biochim Biophys Acta. 2016;1863:2362–78.
Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–46.
Ma Z, Gu S, Song M, Yan C, Hui B, Ji H, et al. Long non-coding RNA SNHG17 is an unfavourable prognostic factor and promotes cell proliferation by epigenetically silencing P57 in colorectal cancer. Mol Biosyst. 2017;13:2350–2361.
Zhang G, Xu Y, Wang S, Gong Z, Zou C, Zhang H, et al. LncRNA SNHG17 promotes gastric cancer progression by epigenetically silencing of p15 and p57. J Cell Physiol. 2019;234:5163–5174.
Xu T, Yan S, Jiang L, Yu S, Lei T, Yang D, et al. Gene amplification-driven long noncoding RNA SNHG17 regulates cell proliferation and migration in human non-small-cell lung cancer. Mol Ther Nucleic Acids. 2019;17:405–13.
Li H, Li T, Huang D, Zhang P. Long noncoding RNA SNHG17 induced by YY1 facilitates the glioma progression through targeting miR-506-3p/CTNNB1 axis to activate Wnt/β-catenin signaling pathway. Cancer Cell Int. 2020;20:29.
Du Y, Wei N, Hong J, Pan W. Long non-coding RNASNHG17 promotes the progression of breast cancer by sponging miR-124-3p. Cancer Cell Int. 2020;20:40.
Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16:421–33.
Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9.
Crea F, Watahiki A, Quagliata L, Xue H, Pikor L, Parolia A, et al. Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget. 2014;5:764–74.
Shiah SG, Hsiao JR, Chang WM, Chen YW, Jin YT, Wong TY, et al. Downregulated miR329 and miR410 promote the proliferation and invasion of oral squamous cell carcinoma by targeting Wnt-7b. Cancer Res. 2014;74:7560–72.
Yu T, Liu K, Wu Y, Fan J, Chen J, Li C, et al. MicroRNA-9 inhibits the proliferation of oral squamous cell carcinoma cells by suppressing expression of CXCR4 via the Wnt/β-catenin signaling pathway. Oncogene. 2014;33:5017–27.
Cui Y, Fang W, Li C, Tang K, Zhang J, Lei Y, et al. Development and validation of a novel signature to predict overall survival in "driver gene-negative" Lung Adenocarcinoma (LUAD): results of a multicenter study. Clin Cancer Res. 2019;25:1546–56.
Wang L, Tang D, Wu T, Sun F. ELF1-mediated LUCAT1 promotes choroidal melanoma by modulating RBX1 expression. Cancer Med. 2020;9:2160–70.
Chen CH, Su LJ, Tsai HT, Hwang CF. ELF-1 expression in nasopharyngeal carcinoma facilitates proliferation and metastasis of cancer cells via modulation of CCL2/CCR2 signaling. Cancer Manag Res. 2019;11:5243–54.
Yang DX, Li NE, Ma Y, Han YC, Shi Y. Expression of Elf-1 and survivin in non-small cell lung cancer and their relationship to intratumoral microvessel density. Chin J Cancer. 2010;29:396–402.
Takai N, Miyazaki T, Nishida M, Shang S, Nasu K, Miyakawa I. Clinical relevance of Elf-1 overexpression in endometrial carcinoma. Gynecol Oncol. 2003;89:408–13.
Budka JA, Ferris MW, Capone MJ, Hollenhorst PC. Common ELF1 deletion in prostate cancer bolsters oncogenic ETS function, inhibits senescence and promotes docetaxel resistance. Genes Cancer. 2018;9:198–214.
Paczkowska J, Soloch N, Bodnar M, Kiwerska K, Janiszewska J, Vogt J, et al. Expression of ELF1, a lymphoid ETS domain-containing transcription factor, is recurrently lost in classical Hodgkin lymphoma. Br J Haematol. 2019;185:79–88.
Acknowledgements
Authors deeply appreciate all participants in this study.
Funding
This study was supported by Natural Science Foundation for the Youth of China (No. 81400488), Jilin Provincial Science and Technology Department (No. 20180520058JH), Jilin Province Health and Health Technology Innovation Project (No. 2018J073).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Qiao, C., Qiao, T., Yang, S. et al. SNHG17/miR-384/ELF1 axis promotes cell growth by transcriptional regulation of CTNNB1 to activate Wnt/β-catenin pathway in oral squamous cell carcinoma. Cancer Gene Ther 29, 122–132 (2022). https://doi.org/10.1038/s41417-021-00294-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-021-00294-9
This article is cited by
-
Novel biomarkers to predict treatment response and prognosis in locally advanced rectal cancer undergoing neoadjuvant chemoradiotherapy
BMC Cancer (2023)
-
Oral squamous cell carcinomas: state of the field and emerging directions
International Journal of Oral Science (2023)
-
Long noncoding RNA SNHG17: a novel molecule in human cancers
Cancer Cell International (2022)
-
SNHG17 promotes colorectal tumorigenesis and metastasis via regulating Trim23-PES1 axis and miR-339-5p-FOSL2-SNHG17 positive feedback loop
Journal of Experimental & Clinical Cancer Research (2021)