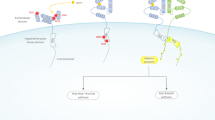
Abstract
OPCML is a highly conserved glycosyl phosphatidylinositol (GPI)-anchored protein belonging to the IgLON family of cell adhesion molecules. OPCML functions as a tumor suppressor and is silenced in over 80% of ovarian cancers by loss of heterozygosity and by epigenetic mechanisms. OPCML inactivation is also observed in many other cancers suggesting a conservation of tumor suppressor function. Although epigenetic silencing and subsequent loss of OPCML expression correlate with poor progression-free and overall patient survival, its mechanism of action is only starting to be fully elucidated. Recent discoveries have demonstrated that OPCML exerts its tumor suppressor effect by inhibiting several cancer hallmark phenotypes in vitro and abrogating tumorigenesis in vivo, by downregulating/inactivating a specific spectrum of Receptor Tyrosine Kinases (RTKs), including EphA2, FGFR1, FGFR3, HER2, HER4, and AXL. This modulation of RTKs can also sensitize ovarian and breast cancers to lapatinib, erlotinib, and anti-AXL therapies. Furthermore, OPCML has also been shown to function in synergy with the tumor suppressor phosphatase PTPRG to inactivate pro-metastatic RTKs such as AXL. Recently, the identification of inactivating point mutations and the elucidation of the crystal structure of OPCML have provided valuable insights into its structure-function relationships, giving rise to its potential as an anti-cancer therapeutic.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Sellar GC, Watt KP, Rabiasz GJ, Stronach EA, Li L, Miller EP, et al. OPCML at 11q25 is epigenetically inactivated and has tumor-suppressor function in epithelial ovarian cancer. Nat Genet. 2003;34:337–43.
McKie AB, Vaughan S, Zanini E, Okon IS, Louis L, de Sousa C, et al. The OPCML tumor suppressor functions as a cell surface repressor-adaptor, negatively regulating receptor tyrosine kinases in epithelial ovarian. Cancer Cancer Discov. 2012;2:156–71.
Antony J, Zanini E, Kelly Z, Tan TZ, Karali E, Alomary M, et al. The tumour suppressor OPCML promotes AXL inactivation by the phosphatase PTPRG in ovarian cancer. EMBO Rep. 2018;19:e45670.
Zanini E, Louis LS, Antony J, Karali E, Okon IS, McKie AB, et al. The tumor suppressor protein OPCML potentiates anti-EGFR and anti-HER2 targeted therapy in HER2-positive ovarian and breast cancer. Mol Cancer Ther. 2017;16:2246–2256.
Birtley JR, Alomary M, Zanini E, Antony J, Maben Z, Weaver GC, et al. Inactivating mutations and X-ray crystal structure of the tumor suppressor OPCML reveal cancer-associated functions. Nat Commun 2019;10:3134.
Cho TM, Hasegawa JI, Ge BL, Loh HH. Purification to apparent homogeneity of a μ-type opioid receptor from rat brain. Proc Natl Acad Sci USA. 1986;83:4138–42.
Schofield PR, McFarland KC, Hayflick JS, Wilcox JN, Cho TM, Roy S, et al. Molecular characterization of a new immunoglobulin superfamily protein with potential roles in opioid binding and cell contact. EMBO J. 1989;8:489–95.
Lodge AP, Howard MR, McNamee CJ, Moss DJ. Co-localisation, heterophilic interactions and regulated expression of IgLON family proteins in the chick nervous system. Mol Brain Res. 2000;82:84–94.
Pimenta AF, Fischer I, Levitt P. cDNA cloning and structural analysis of the human limbic-system-associated membrane protein (LAMP). Gene. 1996;170:189–95.
Gil OD, Zanazzi G, Struyk AF, Salzer JL. Neurotrimin mediates bifunctional effects on neurite outgrowth via homophilic and heterophilic interactions. J Neurosci. 1998;18:9312–25.
Funatsu N, Miyata S, Kumanogoh H, Shigeta M, Hamada K, Endo Y, et al. Characterization of a novel rat brain glycosylphosphatidylinositol-anchored protein (Kilon), a member of the IgLON cell adhesion molecule family. J Biol Chem. 1999;274:8224–30.
Sabater L, Gaig C, Gelpi E, Bataller L, Lewerenz J, Torres-Vega E, et al. A novel non-rapid-eye movement and rapid-eye-movement parasomnia with sleep breathing disorder associated with antibodies to IgLON5: a case series, characterisation of the antigen, and post-mortem study. Lancet Neurol. 2014;13:575–86.
Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–9.
Reed JE, Dunn JR, du Plessis DG, Shaw EJ, Reeves P, Gee A. L. et al. Expression of cellular adhesion molecule “OPCML” is down-regulated in gliomas and other brain tumours. Neuropathol Appl Neurobiol. 2007;33:77–85.
Miyata S, Taguchi K, Maekawa S. Dendrite-associated opioid-binding cell adhesion molecule localizes at neurosecretory granules in the hypothalamic magnocellular neurons. Neuroscience. 2003;122:169–81.
Miyata S, Matsumoto N, Maekawa S. Polarized targeting of IgLON cell adhesion molecule OBCAM to dendrites in cultured neurons. Brain Res. 2003;979:129–36.
Miyata S, Matsumoto N, Taguchi K, Akagi A, Iino T, Funatsu N, et al. Biochemical and ultrastructural analyses of iglon cell adhesion molecules, kilon and obcam in the rat brain. Neuroscience. 2003;117:645–58.
Eagleson KL, Pimenta AF, Burns MM, Fairfull LD, Cornuet PK, Zhang L, et al. Distinct domains of the limbic system-associated membrane protein (LAMP) mediate discrete effects on neurite outgrowth. Mol Cell Neurosci. 2003;24:725–40.
Itoh S, Hachisuka A, Kawasaki N, Hashii N, Teshima R, Hayakawa T, et al. Glycosylation analysis of IgLON family proteins in rat brain by liquid chromatography and multiple-stage mass spectrometry. Biochemistry. 2008;47:10132–54.
Reed J, McNamee C, Rackstraw S, Jenkins J, Moss D. Diglons are heterodimeric proteins composed of IgLON subunits, and Diglon-CO inhibits neurite outgrowth from cerebellar granule cells. J Cell Sci. 2004;117:3961–73.
Gil OD, Zhang L, Chen S, Ren YQ, Pimenta A, Zanazzi G, et al. Complementary expression and heterophilic interactions between igLON family members neurotrimin and LAMP. J Neurobiol. 2002;51:190–204.
Ranaivoson FM, Turk LS, Ozgul S, Kakehi S, Daake S, von, Lopez N, et al. A proteomic screen of neuronal cell-surface molecules reveals IgLONs as structurally conserved interaction modules at the synapse. Structure. 2019;27:893–906.
Ntougkos E, Rush R, Scott D, Frankenberg T, Gabra H, Smyth JF, et al. The IgLON family in epithelial ovarian cancer: expression profiles and clinicopathologic correlates. Clin Cancer Res. 2005;11:5764–8.
Chen J, Lui W-O, Vos MD, Clark GJ, Takahashi M, Schoumans J, et al. The t(1;3) breakpoint-spanning genes LSAMP and NORE1 are involved in clear cell renal cell carcinomas. Cancer Cell. 2003;4:405–13.
Kresse SH, Ohnstad HO, Paulsen EB, Bjerkehagen B, Szuhai K. LSAMP, a novel candidate tumor suppressor gene in human osteosarcomas, identified by array comparative genomic hybridization. Genes Chromosomes Cancer. 2009;693:679–93.
Zhao J, Bradfield JP, Li M, Wang K, Zhang H, Kim CE, et al. The role of obesity-associated loci identified in genome-wide association studies in the determination of pediatric BMI. Obesity. 2009;17:2254–7.
Takita J, Chen Y, Okubo J, Sanada M, Adachi M, Ohki K, et al. Aberrations of NEGR1 on 1p31 and MYEOV on 11q13 in neuroblastoma. Cancer Sci. 2011;102:1645–50.
Ye M, Parente F, Li X, Perryman MB, Zelante L, Wynshaw-Boris A, et al. Gene-targeted deletion of OPCML and Neurotrimin in mice does not yield congenital heart defects. Am J Med Genet Part A. 2014;164:966–74.
Gabra H, Taylor L, Cohen B, Lessels A, Eccles D, Leonard R, et al. Chromosome 11 allele imbalance and clinicopathological correlates in ovarian tumours. Br J Cancer. 1995;72:367–75.
Thiagalingam S, Foy RL, Cheng K, Lee HJ, Thiagalingam A, Ponte JF. Loss of heterozygosity as a predictor to map tumor suppressor genes in cancer: molecular basis of its occurrence. Curr Opin Oncol. 2002;14:65–72.
Stronach EA, Sellar GC, Blenkiron C, Rabiasz GJ, Taylor KJ, Miller EP, et al. Identification of clinically relevant genes on chromosome 11 in a functional model of ovarian cancer tumor suppression. Cancer Res. 2003;63:8648–55.
Czekierdowski A, S C, M S, M W, P K, J. K. Opioid-binding protein/cell adhesion molecule-like (OPCML) gene and promoter methylation status in women with ovarian cancer. Neuroendocrinol. 2006;27:609–13.
Ye F, Zhang S-F, Xie X, Lu W-G. OPCML gene promoter methylation and gene expression in tumor and stroma cells of invasive cervical carcinoma. Cancer Invest. 2008;26:569–74.
Anglim PP, Galler JS, Koss MN, Hagen JA, Turla S, Campan M, et al. Identification of a panel of sensitive and specific DNA methylation markers for squamous cell lung cancer. Mol Cancer. 2008;7:62.
Mei FC, Young TW, Liu J, Cheng X. RAS-mediated epigenetic inactivation of OPCML in oncogenic transformation of human ovarian surface epithelial cells. FASEB J. 2006;20:497–9.
Duarte-Pereira S, Paiva F, Costa VL, Ramalho-Carvalho J, Savva-Bordalo J, Rodrigues A, et al. Prognostic value of opioid binding protein/cell adhesion molecule-like promoter methylation in bladder carcinoma. Eur J Cancer. 2011;47:1106–14.
Wu Y, Davison J, Qu X, Morrissey C, Storer B, Brown L, et al. Methylation profiling identified novel differentially methylated markers including OPCML and FLRT2 in prostate cancer. Epigenetics. 2016;11:247–58.
Xing X, Cai W, Ma S, Wang Y, Shi H, Li M, et al. Down-regulated expression of OPCML predicts an unfavorable prognosis and promotes disease progression in human gastric cancer. BMC Cancer. 2017;17:268.
Cui Y, Ying Y, van Hasselt A, Ng KM, Yu J, Zhang Q, et al. OPCML is a broad tumor suppressor for multiple carcinomas and lymphomas with frequently epigenetic inactivation. PLoS ONE. 2008;3:e2990.
McNamee CJ, Reed JE, Howard MR, Lodge AP, Moss DJ. Promotion of neuronal cell adhesion by members of the IgLON family occurs in the absence of either support or modification of neurite outgrowth. J Neurochem. 2002;80:941–8.
Pike LJ. Growth factor receptors, lipid rafts and caveolae: an evolving story. Biochim Biophys Acta. 2005;1746:260–73.
Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–72.
Tzahar E, Waterman H, Chen X, Levkowitz G, Karunagaran D, Lavi S, et al. A hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Mol Cell Biol. 1996;16:5276–87.
Graus-Porta D, Beerli RR, Daly JM, Hynes NE. ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J. 1997;16:1647–55.
Geyer CE, Forster J, Lindquist D, Chan S, Romieu CG, Pienkowski T, et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N. Engl J Med. 2006;355:2733–43.
Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, et al. Erlotinib in previously treated non–small-cell lung cancer. N. Engl J Med. 2005;353:123–32.
Antony J, Huang RYJ, AXL-driven EMT. state as a targetable conduit in cancer. Cancer Res. 2017;77:3725–32.
Huang RY-J, Antony J, Tan TZ, Tan DS-P. Targeting the AXL signaling pathway in ovarian cancer. Mol Cell Oncol. 2017;4:e1263716.
Antony J, Tan TZ, Kelly Z, Low J, Choolani M, Recchi C, et al. The GAS6-AXL signaling network is a mesenchymal (Mes) molecular subtype-specific therapeutic target for ovarian cancer. Sci Signal. 2016;9:ra97–ra97.
Gjerdrum C, Tiron C, Hoiby T, Stefansson I, Haugen H, Sandal T, et al. Axl is an essential epithelial-to-mesenchymal transition-induced regulator of breast cancer metastasis and patient survival. Proc Natl Acad Sci USA. 2010;107:1124–9.
Elkabets M, Pazarentzos E, Juric D, Sheng Q, Pelossof RA, Brook S, et al. AXL mediates resistance to pi3kα inhibition by activating the EGFR/PKC/mTOR axis in head and neck and esophageal squamous cell carcinomas. Cancer Cell. 2015;27:533–46.
Byers LA, Diao L, Wang J, Saintigny P, Girard L, Peyton M, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res. 2013;19:279–90.
Sasaki T, Knyazev PG, Clout NJ, Cheburkin Y, Göhring W, Ullrich A, et al. Structural basis for Gas6-Axl signalling. EMBO J. 2006;25:80–7.
Shu ST, Sugimoto Y, Liu S, Chang HL, Ye W, Wang LS, et al. Function and regulatory mechanisms of the candidate tumor suppressor receptor Protein Tyrosine Phosphatase Gamma (PTPRG) in breast cancer cells. Anticancer Res. 2010;30:1937–46.
Nieto MA, Huang RY-J, Jackson RA, Thiery JP. EMT: 2016. Cell 2016;166:21–45.
Thiery JP, Lim CT. Tumor dissemination: an EMT affair. Cancer Cell 2013;23:272–3.
Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell 2009;139:871–90.
Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–54.
Thiery JP, Sleeman JP. Complex networks orchestrate epithelial–mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–42.
Zurzolo, C. Synergistic inactivation of AXL: a (cross)road to cure ovarian cancer? EMBO Rep. 2018;19:e46492.
Holland SJ, Pan A, Franci C, Hu Y, Chang B, Li W, et al. R428, a selective small molecule inhibitor of Axl kinase, blocks tumor spread and prolongs survival in models of metastatic breast cancer. Cancer Res. 2010;70:1544–54.
Li C, Tang L, Zhao L, Li L, Xiao Q, Luo X, et al. OPCML is frequently methylated in human colorectal cancer and its restored expression reverses EMT via downregulation of smad signaling. Am J Cancer Res. 2015;5:1635–48.
Brown KA, Ham A-JL, Clark CN, Meller N, Law BK, Chytil A, et al. Identification of novel Smad2 and Smad3 associated proteins in response to TGF-β1. J Cell Biochem. 2008;105:596–611.
Wang LH, Chu FF, Ren DH, Du X. Effect of OPCML gene on the biological behavior of gastric cancer cell line AGS. J Biol Regul Homeost Agents. 2016;30:529–34.
Wu SY, Sood AK. New roles opined for OPCML. Cancer Discov. 2012;2:115–6.
Acknowledgements
JA, EZ, JRB, HG, and CR received no financial assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
HG has ownership interest (including patents) to develop OPCML-based therapeutics. He is also Chief Medical Officer at BergenBio (producer of the AXL inhibitor BGB324). The remaining authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Antony, J., Zanini, E., Birtley, J.R. et al. Emerging roles for the GPI-anchored tumor suppressor OPCML in cancers. Cancer Gene Ther 28, 18–26 (2021). https://doi.org/10.1038/s41417-020-0187-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-020-0187-6
This article is cited by
-
Human chromosome 3p21.3 carries TERT transcriptional regulators in pancreatic cancer
Scientific Reports (2021)