Abstract
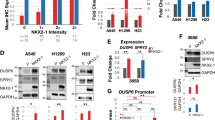
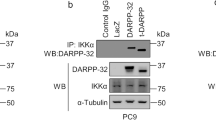
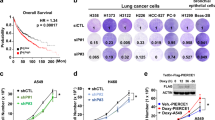
KRAS is one of the most frequently mutated oncogenes, especially in lung cancers. Targeting of KRAS directly or the downstream effector signaling machinery is of prime interest in treating lung cancers. Here, we uncover that ERK3, a ubiquitously expressed atypical MAPK, is required for KRAS-mediated NSCLC tumors. ERK3 is highly expressed in lung cancers, and oncogenic KRAS led to the activation and stabilization of the ERK3 protein. In particular, phosphorylation of serine 189 in the activation motif of ERK3 is significantly increased in lung adenocarcinomas in comparison to adjacent normal controls in patients. Loss of ERK3 prevents the anchorage-independent growth of KRAS G12C-transformed human bronchial epithelial cells. We further find that loss of ERK3 reduces the oncogenic growth of KRAS G12C-driven NSCLC tumors in vivo and that the kinase activity of ERK3 is required for KRAS-driven oncogenesis in vitro. Our results demonstrate an obligatory role for ERK3 in NSCLC tumor progression and suggest that ERK3 kinase inhibitors can be pursued for treating KRAS G12C-driven tumors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
30 October 2020
This Article was originally published under an Open Access licence [CC BY 4.0] in error. It should have been published under Nature Research’s License to Publish. The PDF and HTML versions of the Article have been corrected accordingly.
09 November 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41417-020-00250-z
References
Cheng TY, Cramb SM, Baade PD, Youlden DR, Nwogu C, Reid ME. The international epidemiology of lung cancer: latest trends, disparities, and tumor characteristics. J Thorac Oncol. 2016;11:1653–71.
Pikor LA, Ramnarine VR, Lam S, Lam WL. Genetic alterations defining NSCLC subtypes and their therapeutic implications. Lung Cancer. 2013;82:179–89.
Yuan M, Huang LL, Chen JH, Wu J, Xu Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct Target Ther. 2019;4:61.
Liu TC, Jin X, Wang Y, Wang K. Role of epidermal growth factor receptor in lung cancer and targeted therapies. Am J Cancer Res. 2017;7:187–202.
Califano R, Abidin A, Tariq NU, Economopoulou P, Metro G, Mountzios G. Beyond EGFR and ALK inhibition: unravelling and exploiting novel genetic alterations in advanced non small-cell lung cancer. Cancer Treat Rev. 2015;41:401–11.
Yang S, Yu X, Fan Y, Shi X, Jin Y. Clinicopathologic characteristics and survival outcome in patients with advanced lung adenocarcinoma and KRAS mutation. J Cancer. 2018;9:2930–7.
Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–90.
Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–30.
Mitin N, Rossman KL, Der CJ. Signaling interplay in Ras superfamily function. Curr Biol. 2005;15:R563–574.
Pradhan R, Singhvi G, Dubey SK, Gupta G, Dua K. MAPK pathway: a potential target for the treatment of non-small-cell lung carcinoma. Future Med Chem. 2019;11:793–5.
Raman M, Chen W, Cobb MH. Differential regulation and properties of MAPKs. Oncogene. 2007;26:3100–12.
Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol. 2004;5:875–85.
Hymowitz SG, Malek S. Targeting the MAPK pathway in RAS mutant cancers. Cold Spring Harb Perspect Med. 2018;8:a031492.
Braicu C, Buse M, Busuioc C, Drula R, Gulei D, Raduly L. et al. A comprehensive review on MAPK: a promising therapeutic target in cancer. Cancers. 2019;11:1618.
Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev: MMBR. 2011;75:50–83.
Klinger S, Turgeon B, Levesque K, Wood GA, Aagaard-Tillery KM, Meloche S. Loss of Erk3 function in mice leads to intrauterine growth restriction, pulmonary immaturity, and neonatal lethality. Proc Natl Acad Sci USA. 2009;106:16710–5.
Ronkina N, Schuster-Gossler K, Hansmann F, Kunze-Schumacher H, Sandrock I, Yakovleva T. et al. Germ line deletion reveals a nonessential role of atypical mitogen-activated protein kinase 6/extracellular signal-regulated kinase 3. Mol Cell Biol. 2019;39:1–11.
Soulez M, Saba-El-Leil MK, Turgeon B, Mathien S, Coulombe P, Klinger S. et al. Reevaluation of the role of extracellular signal-regulated kinase 3 in perinatal survival and postnatal growth using new genetically engineered mouse models. Mol Cell Biol. 2019;39:1–10.
Coulombe P, Rodier G, Bonneil E, Thibault P, Meloche S. N-Terminal ubiquitination of extracellular signal-regulated kinase 3 and p21 directs their degradation by the proteasome. Mol Cell Biol. 2004;24:6140–50.
Al-Mahdi R, Babteen N, Thillai K, Holt M, Johansen B, Wetting HL, et al. A novel role for atypical MAPK kinase ERK3 in regulating breast cancer cell morphology and migration. Cell Adhes Migr. 2015;9:483–94.
Elkhadragy L, Alsaran H, Morel M, Long W. Activation loop phosphorylation of ERK3 is important for its kinase activity and ability to promote lung cancer cell invasiveness. J Biol Chem. 2018;293:16193–205.
Long W, Foulds CE, Qin J, Liu J, Ding C, Lonard DM, et al. ERK3 signals through SRC-3 coactivator to promote human lung cancer cell invasion. J Clin Investig. 2012;122:1869–80.
Bogucka K, Pompaiah M, Marini F, Binder H, Harms G, Kaulich M. et al. ERK3/MAPK6 controls IL-8 production and chemotaxis. eLife. 2020;9:e52511.
Dearden S, Stevens J, Wu YL, Blowers D. Mutation incidence and coincidence in non small-cell lung cancer: meta-analyses by ethnicity and histology (mutMap). Ann Oncol. 2013;24:2371–6.
Katz M, Amit I, Yarden Y. Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochimica et biophysica acta. 2007;1773:1161–76.
Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–67.
Fukazawa H, Mizuno S, Uehara Y. A microplate assay for quantitation of anchorage-independent growth of transformed cells. Anal Biochem. 1995;228:83–90.
Alley MC, Scudiero DA, Monks A, Hursey ML, Czerwinski MJ, Fine DL, et al. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res. 1988;48:589–601.
Alley MC, Uhl CB, Lieber MM. Improved detection of drug cytotoxicity in the soft agar colony formation assay through use of a metabolizable tetrazolium salt. Life Sci. 1982;31:3071–8.
Anderson SN, Towne DL, Burns DJ, Warrior U. A high-throughput soft agar assay for identification of anticancer compound. J Biomol Screen. 2007;12:938–45.
Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, et al. Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell. 2018;172:578–89 e517.
Diaz R, Nguewa PA, Parrondo R, Perez-Stable C, Manrique I, Redrado M, et al. Antitumor and antiangiogenic effect of the dual EGFR and HER-2 tyrosine kinase inhibitor lapatinib in a lung cancer model. BMC Cancer. 2010;10:188.
Mooz J, Oberoi-Khanuja TK, Harms GS, Wang W, Jaiswal BS, Seshagiri S, et al. Dimerization of the kinase ARAF promotes MAPK pathway activation and cell migration. Sci Signal. 2014;7:ra73.
Lundberg AS, Randell SH, Stewart SA, Elenbaas B, Hartwell KA, Brooks MW, et al. Immortalization and transformation of primary human airway epithelial cells by gene transfer. Oncogene. 2002;21:4577–86.
Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–91.
Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–4.
Ke N, Albers A, Claassen G, Yu DH, Chatterton JE, Hu X, et al. One-week 96-well soft agar growth assay for cancer target validation. Biotechniques. 2004;36:826–8.
McCarty KS Jr., Miller LS, Cox EB, Konrath J, McCarty KS Sr. Estrogen receptor analyses. Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Arch Pathol Lab Med. 1985;109:716–21.
Mertins P, Tang LC, Krug K, Clark DJ, Gritsenko MA, Chen L, et al. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat Protoc. 2018;13:1632–61.
Chen F, Zhang Y, Gibbons DL, Deneen B, Kwiatkowski DJ, Ittmann M, et al. Pan-cancer molecular classes transcending tumor lineage across 32 cancer types, multiple data platforms, and over 10,000 cases. Clin Cancer Res. 2018;24:2182–93.
Acknowledgements
We thank Stefanie Wenzel for excellent technical assistance. This work is supported from the grants from Else Kroener Fresenius Stiftung; MERCK (project ID-ERK-KR), CRC 1292, TP12, and Forschungszentrum für Immuntherapie of the University Medical Center Mainz to KR. We thank Dr. Daniela Hoeller for critical reading of the paper. We thank Dr. Bernd Gromol for his help with the H scoring and Dr. Jonathan Woodsmith for help with the analysis of Omics data, KR is supported through a Heisenberg professorship of the DFG (RA1739/4-1). We would like to thank the Translational oncology team of MERCK for their valuable inputs and advise. We thank Dr. Christiane Schoenfeld for analyses of the animal experiments.
Author information
Authors and Affiliations
Contributions
KB designed and performed most of the experiments as well as analyzed/interpreted the data prepared Figures and wrote the paper. FM performed Bioinformatic analyses. KD contributed to the staining and analyses of NSCLC tissue microarray. JS, HJ, and SR contributed to xenografts experiments. MS contributed to the bioinformatic analysis of RNA sequencing. KR contributed to the conception, design, analyzed/interpreted the data, and supervised the study. KB and KR wrote the paper with input from all authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Bogucka, K., Marini, F., Rosigkeit, S. et al. ERK3/MAPK6 is required for KRAS-mediated NSCLC tumorigenesis. Cancer Gene Ther 28, 359–374 (2021). https://doi.org/10.1038/s41417-020-00245-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-020-00245-w
This article is cited by
-
Inactivation of EGLN3 hydroxylase facilitates Erk3 degradation via autophagy and impedes lung cancer growth
Oncogene (2022)
-
KRAS mutation: from undruggable to druggable in cancer
Signal Transduction and Targeted Therapy (2021)