Abstract
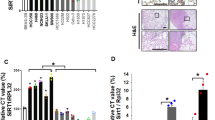
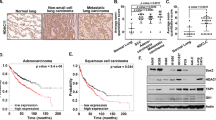
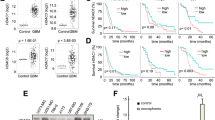
Mutations in the gene encoding for the histone acetyltransferase (HAT) CREBBP are common driver events in multiple types of human cancer, such as small cell lung cancer (SCLC) or Sonic Hedgehog medulloblastoma (SHH MB). Therefore, therapeutic options targeting such alterations are highly desired. We used human cell lines from SCLC as well as primary mouse tumor cells and genetically engineered mouse models for SHH MB to test treatment options with histone deacetylase inhibitors (HDACi) in CREBBP wild-type and mutated tumors. In contrast to CREBBP wild-type SCLC cells, CREBBP-mutated SCLC cells showed significantly lower IC50 values after treatment with HDACi. In addition, both in vitro and in vivo, HDACi had significant effects on cell proliferation of SHH-driven tumor MB cells harboring a CREBBP-mutation as compared to CREBBP wild-type controls. These data suggest that HDACi may serve as an additional therapeutic option for patients suffering from tumors driven by CREBBP mutations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–5. https://doi.org/10.1038/nature11284
Northcott PA, Buchhalter I, Morrissy AS, Hovestadt V, Weischenfeldt J, Ehrenberger T, et al. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547:311–7. https://doi.org/10.1038/nature22973
Kool M, Jones DT, Jager N, Northcott PA, Pugh TJ, Hovestadt V, et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell. 2014;25:393–405. https://doi.org/10.1016/j.ccr.2014.02.004
Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–10. https://doi.org/10.1038/ng.2396
Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471:235–9. https://doi.org/10.1038/nature09727
Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–95. https://doi.org/10.1038/nature09730
Taylor MD, Mainprize TG, Rutka JT, Becker L, Bayani J, Drake JM. Medulloblastoma in a child with Rubenstein-Taybi Syndrome: case report and review of the literature. Pedia Neurosurg. 2001;35:235–8. https://doi.org/10.1159/000050428
Miller RW, Rubinstein JH. Tumors in Rubinstein-Taybi syndrome. Am J Med Genet. 1995;56:112–5. https://doi.org/10.1002/ajmg.1320560125
Merk DJ, Ohli J, Merk ND, Thatikonda V, Morrissy S, Schoof M, et al. Opposing effects of CREBBP mutations govern the phenotype of Rubinstein-Taybi syndrome and adult SHH medulloblastoma. Dev Cell. 2018;44:709–.e706. https://doi.org/10.1016/j.devcel.2018.02.012
Mao J, Ligon KL, Rakhlin EY, Thayer SP, Bronson RT, Rowitch D, et al. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res. 2006;66:10171–8. https://doi.org/10.1158/0008-5472.Can-06-0657
Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. https://doi.org/10.1016/j.neuron.2005.08.028
Zhang Z, Hofmann C, Casanova E, Schutz G, Lutz B. Generation of a conditional allele of the CBP gene in mouse. Genesis. 2004;40:82–89. https://doi.org/10.1002/gene.20068
Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–9.
Pei Y, Liu KW, Wang J, Garancher A, Tao R, Esparza LA, et al. HDAC and PI3K antagonists cooperate to inhibit growth of MYC-driven medulloblastoma. Cancer Cell. 2016;29:311–23. https://doi.org/10.1016/j.ccell.2016.02.011
Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14:123–34. https://doi.org/10.1016/j.ccr.2008.07.005
Karpova NN. Role of BDNF epigenetics in activity-dependent neuronal plasticity. Neuropharmacology. 2014;76(Pt C):709–18. https://doi.org/10.1016/j.neuropharm.2013.04.002
Koppel I, Timmusk T. Differential regulation of Bdnf expression in cortical neurons by class-selective histone deacetylase inhibitors. Neuropharmacology. 2013;75:106–15. https://doi.org/10.1016/j.neuropharm.2013.07.015
Wu X, Chen PS, Dallas S, Wilson B, Block ML, Wang CC, et al. Histone deacetylase inhibitors up-regulate astrocyte GDNF and BDNF gene transcription and protect dopaminergic neurons. Int J Neuropsychopharmacol. 2008;11:1123–34. https://doi.org/10.1017/s1461145708009024
Jia D, Augert A, Kim DW, Eastwood E, Wu N, Ibrahim AH, et al. Crebbp loss drives small cell lung cancer and increases sensitivity to HDAC inhibition. Cancer Discov. 2018;8:1422–37. https://doi.org/10.1158/2159-8290.Cd-18-0385
Acknowledgements
We are indebted to Michael Schmidt, Silvia Occhionero, Marie-Christin Burmester. and Veronika Kaltenbrunn for excellent technical support.
Funding
This work was supported by grants from the German Cancer Aid, the Wilhelm-Sander Stiftung, and the Fördergemeinschaft Kinderkrebs-Zentrum Hamburg.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hellwig, M., Merk, D.J., Lutz, B. et al. Preferential sensitivity to HDAC inhibitors in tumors with CREBBP mutation. Cancer Gene Ther 27, 294–300 (2020). https://doi.org/10.1038/s41417-019-0099-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-019-0099-5
This article is cited by
-
Genomic and immunological profiles of small-cell lung cancer between East Asians and Caucasian
Cancer Cell International (2022)
-
Comparison of the mutational profiles of neuroendocrine breast tumours, invasive ductal carcinomas and pancreatic neuroendocrine carcinomas
Oncogenesis (2022)
-
Advances in the Treatment of Hairy Cell Leukemia Variant
Current Treatment Options in Oncology (2022)