Abstract
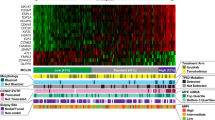
To gain insight into the molecular pathogenesis of patients with mantle cell lymphoma (MCL), next-generation whole-exome sequencing of 16 MCL patients was performed. We identified recurrent mutations in genes that are well known to be functionally relevant in MCL, including ATM (37.5%), TP53 (31.3%), WHSC1 (31.3%), CCND1 (18.8%), NOTCH2 (6.3%), and CDKN2A (6.3%). We also identified somatic mutations in genes for which a functional role in MCL has not been previously suspected. These genes included CCDC15, APC, CDH1, S1PR1, ATRX, BRCA2, CASP8, and NOTCH3. Further, we investigated the prognostic factors associated with MCL from clinical, pathological, and genetic mutations. Mutations of TP53 (P = 0.021) was a significant prognostic factor with shorter overall survival (OS). Although there was no statistical difference, the median survival time of patients with WHSC1 mutations was shorter than those without mutations (P = 0.070). Mutations in ATM and CCND1 had no prognostic value (P = 0.552, 0.566). When adjusted for MCL International Prognostic Index (MIPI) or combined MCL-International Prognostic Index (MIPI-c), TP53 and WHSC1 mutations were the most important prognostic factors in MCL (P < 0.05). Our data provide an unbiased view of the landscape of mutations in MCL and commend that all patients benefit from mutations of TP53 and WHSC1 at diagnosis, in addition to MIPI and MIPI-c score.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ghielmini M, Zucca E. How I treat mantle cell lymphoma. Blood. 2009;114:1469–76.
Jares P, Colomer D, Campo E. Genetic and molecular pathogenesis of mantle cell lymphoma: perspectives for new targeted therapeutics. Nat Rev Cancer. 2007;7:750–62.
Leux C, Maynadie M, Troussard X, Cabrera Q, Herry A, Le Guyader-Peyrou S, et al. Mantle cell lymphoma epidemiology: a population-based study in France. Ann Hematol. 2014;93:1327–33.
Zhou Y, Wang H, Fang W, Romaguer JE, Zhang Y, Delasalle KB, et al. Incidence trends of mantle cell lymphoma in the United States between 1992 and 2004. Cancer. 2008;113:791–8.
Cheah CY, Seymour JF, Wang ML. Mantle cell lymphoma. J Clin Oncol. 2016;34:1256–69.
Vose JM. Mantle cell lymphoma: 2012 update on diagnosis, risk-stratification, and clinical management. Am J Hematol. 2012;87:604–9.
Jares P, Colomer D, Campo E. Molecular pathogenesis of mantle cell lymphoma. J Clin Invest. 2012;122:3416–23.
Jares P, Campo E. Advances in the understanding of mantle cell lymphoma. Br J Haematol. 2008;142:149–65.
Inamdar AA, Goy A, Ayoub NM, Attia C, Oton L, Taruvai V, et al. Mantle cell lymphoma in the era of precision medicine-diagnosis, biomarkers and therapeutic agents. Oncotarget. 2016;7:48692–731.
Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci USA. 2012;109:3879–84.
Miranda NF, Georgiou K, Chen L, Wu C, Gao Z, Zaravinos A, et al. Exome sequencing reveals novel mutation targets in diffuse large B-cell lymphomas derived from Chinese patients. Blood. 2014;124:2544–53.
Cheson BD, Horning SJ, Coiffier B, Shipp MA, Fisher RI, Connors JM, et al. Report of an international workshop to standardize response criteria for non-Hodgkin’s lymphomas. NCI Sponsored International Working Group. J Clin Oncol. 1999;17:244.
Hoster E, Dreyling M, Klapper W, Gisselbrecht C, van Hoof A, Kluin-Nelemans HC, et al. A new prognostic index (MIPI) for patients with advanced-stage mantle cell lymphoma. Blood. 2008;111:558–65.
Hoster E, Rosenwald A, Berger F, Bernd HW, Hartmann S, Loddenkemper C, et al. Prognostic value of Ki-67 index, cytology, and growth pattern in mantle-cell lymphoma: results from randomized trials of the European Mantle Cell Lymphoma Network. J Clin Oncol. 2016;34:1386–94.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinforma. 2013;43:11. 10 11-33
Li H, Durbin R. Fast and accurate long-read alignment with Burrows−Wheeler transform. Bioinformatics. 2010;26:589–95.
Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–76.
Fabregat A, Sidiropoulos K, Viteri G, Forner O, Marin-Garcia P, Arnau V, et al. Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinforma. 2017;18:142.
Ahmed M, Zhang L, Nomie K, Lam L, Wang M. Gene mutations and actionable genetic lesions in mantle cell lymphoma. Oncotarget. 2016;7:58638–48.
Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82.
Oyer JA, Huang X, Zheng Y, Shim J, Ezponda T, Carpenter Z, et al. Point mutation E1099K in MMSET/NSD2 enhances its methyltranferase activity and leads to altered global chromatin methylation in lymphoid malignancies. Leukemia. 2014;28:198–201.
Bea S, Valdes-Mas R, Navarro A, Salaverria I, Martin-Garcia D, Jares P, et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci USA. 2013;110:18250–5.
Zhang J, Jima D, Moffitt AB, Liu Q, Czader M, Hsi ED, et al. The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells. Blood. 2014;123:2988–96.
Tamaki H, Sanda M, Katsumata O, Hara Y, Fukaya M, Sakagami H. Pilt is a coiled-coil domain-containing protein that localizes at the trans-Golgi complex and regulates its structure. FEBS Lett. 2012;586:3064–70.
Morra F, Luise C, Visconti R, Staibano S, Merolla F, Ilardi G, et al. New therapeutic perspectives in CCDC6 deficient lung cancer cells. Int J Cancer. 2015;136:2146–57.
Chen M, Ni J, Chang HC, Lin CY, Muyan M, Yeh S. CCDC62/ERAP75 functions as a coactivator to enhance estrogen receptor beta-mediated transactivation and target gene expression in prostate cancer cells. Carcinogenesis. 2009;30:841–50.
Spiegel S, Milstien S. Sphingosine 1-phosphate, a key cell signaling molecule. J Biol Chem. 2002;277:25851–4.
Selvam SP, Ogretmen B. Sphingosine kinase/sphingosine 1-phosphate signaling in cancer therapeutics and drug resistance. Handb Exp Pharmacol. 2013;216:3–27.
Bouska A, Zhang W, Gong Q, Iqbal J, Scuto A, Vose J, et al. Combined copy number and mutation analysis identifies oncogenic pathways associated with transformation of follicular lymphoma. Leukemia. 2017;31:83–91.
Wu C, de Miranda NF, Chen L, Wasik AM, Mansouri L, Jurczak W, et al. Genetic heterogeneity in primary and relapsed mantle cell lymphomas: impact of recurrent CARD11 mutations. Oncotarget. 2016;7:38180–90.
Paul Y, Mondal B, Patil V, Somasundaram K. DNA methylation signatures for 2016 WHO classification subtypes of diffuse gliomas. Clin Epigenetics. 2017;9:32.
Herbaux C, Duployez N, Badens C, Poret N, Gardin C, Decamp M, et al. Incidence of ATRX mutations in myelodysplastic syndromes, the value of microcytosis. Am J Hematol. 2015;90:737–8.
Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips KA, Mooij TM, Roos-Blom MJ, Jervis S, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317:2402–16.
Molina-Montes E, Perez-Nevot B, Pollan M, Sanchez-Cantalejo E, Espin J, Sanchez MJ. Cumulative risk of second primary contralateral breast cancer in BRCA1/BRCA2 mutation carriers with a first breast cancer: a systematic review and meta-analysis. Breast. 2014;23:721–42.
Friedenson B. The BRCA1/2 pathway prevents hematologic cancers in addition to breast and ovarian cancers. BMC Cancer. 2007;7:152.
Halldorsdottir AM, Lundin A, Murray F, Mansouri L, Knuutila S, Sundstrom C, et al. Impact of TP53 mutation and 17p deletion in mantle cell lymphoma. Leukemia. 2011;25:1904–8.
Dong HJ, Zhou LT, Fang C, Fan L, Zhu DX, Wang YH, et al. TP53 mutation is not an independent prognostic factor in patients with mantle cell lymphoma at advanced stage. Med Oncol. 2012;29:2166–73.
Nordstrom L, Sernbo S, Eden P, Gronbaek K, Kolstad A, Raty R, et al. SOX11 and TP53 add prognostic information to MIPI in a homogenously treated cohort of mantle cell lymphoma—a Nordic Lymphoma Group study. Br J Haematol. 2014;166:98–108.
Delfau-Larue MH, Klapper W, Berger F, Jardin F, Briere J, Salles G, et al. High-dose cytarabine does not overcome the adverse prognostic value of CDKN2A and TP53 deletions in mantle cell lymphoma. Blood. 2015;126:604–11.
Acknowledgements
The authors thank the patients and their families who contributed to this study. The authors would also like to acknowledge Professor Cuiling Liu in Department of Pathology, Peking University Third Hospital.
Funding
This work was funded by China Health Promotion Foundation, CHPF-zlkysx-001. Employees of the funding source were involved in the collection and assembly of data, genetic sequencing, performing statistical analysis, analyzing and interpreting data, and drafting, reviewing, and approving the manuscript, as reflected in the author contributions statement.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Yang, P., Zhang, W., Wang, J. et al. Genomic landscape and prognostic analysis of mantle cell lymphoma. Cancer Gene Ther 25, 129–140 (2018). https://doi.org/10.1038/s41417-018-0022-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-018-0022-5
This article is cited by
-
Genetic and prognostic analysis of blastoid and pleomorphic mantle cell lymphoma: a multicenter analysis in China
Annals of Hematology (2024)
-
Beyond Bruton’s tyrosine kinase inhibitors in mantle cell lymphoma: bispecific antibodies, antibody–drug conjugates, CAR T-cells, and novel agents
Journal of Hematology & Oncology (2023)
-
A sumoylation program is essential for maintaining the mitotic fidelity in proliferating mantle cell lymphoma cells
Experimental Hematology & Oncology (2022)
-
Complex genetic and histopathological study of 15 patient-derived xenografts of aggressive lymphomas
Laboratory Investigation (2022)
-
Progress in molecular feature of smoldering mantle cell lymphoma
Experimental Hematology & Oncology (2021)