Abstract
Metabolic reprogramming is involved in the pathogenesis of not only cancers but also neurodegenerative diseases, cardiovascular diseases, and infectious diseases. With the progress of metabonomics and proteomics, metabolites have been found to affect protein acylations through providing acyl groups or changing the activities of acyltransferases or deacylases. Reciprocally, protein acylation is involved in key cellular processes relevant to physiology and diseases, such as protein stability, protein subcellular localization, enzyme activity, transcriptional activity, protein–protein interactions and protein–DNA interactions. Herein, we summarize the functional diversity and mechanisms of eight kinds of nonhistone protein acylations in the physiological processes and progression of several diseases. We also highlight the recent progress in the development of inhibitors for acyltransferase, deacylase, and acylation reader proteins for their potential applications in drug discovery.
Similar content being viewed by others
Introduction
Protein post-translational modifications (PTMs) increase the functional diversity of the proteome by the covalent addition of functional groups to proteins. Several PTMs use metabolic intermediates, modifying the epigenetic landscape, the cell signaling networks and providing elegant mechanisms to precisely govern protein function. In the early 1960s, histone acetylation was first discovered to regulate gene transcription.1,2 Since the first nonhistone protein p53 was found to be regulated by acetylation in the 1980s, thousands of nonhistone proteins have been identified as acylation targets. In 2009, a research group at the University of Chicago developed a powerful algorithm named PTMap to identify all possible PTMs with high confidence.3 Since then, the group has taken the lead and reported eight novel acylation modifications on histone lysine (K) residue by using mass spectrometry and biochemistry technologies.4 These modifications include crotonylation,5 malonylation, succinylation,6,7 glutarylation,8 β-hydroxybutyrylation,9 dihydroxyisobutyrylation,10 benzoylation,11 and lactylation.12 In addition to modifications by short-chain acylation, modifications by long-chain fatty acid acylation, such as myristoylation, palmitoylation, and prenylation were also identified in nonhistone proteins in the 1980s.13
The cellular intrinsic metabolic reprogramming and the extrinsic metabolic status in the microenvironment are involved in the regulation of protein acylation. Direct participation of central metabolites in PTMs enables cells to integrate information from metabolism into complex cellular decisions that ensures proper regulation of cellular processes, such as protein stability, protein subcellular localization, enzyme activity, transcriptional activity, protein–protein interactions (PPIs) and protein–DNA interactions. In this review, we provide an overview of the expanding landscape of nonhistone protein acylation, mainly including acetylation, succinylaton, malonylation, crotonylation, β-hydroxybutyrylation, lactylation, myristoylation and palmitoylation (Fig. 1). We discuss the generation of the acyl group donor acyl-CoA, the enzymatic regulation of acylation, how the acylation marks are identified by the reader proteins, as well as the general biological function of protein acylation. Abnormal and imbalanced acylation of nonhistone protein in association with various human diseases, especially cancer will be discussed. We also provide a glimpse into the potential value of protein acylation from a therapeutic view and conclude with a discussion of key open questions and future perspectives.
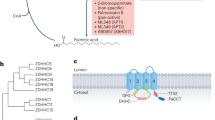
Timeline of the historical milestone for the discovery of protein acylation, and the chemical structures of acyl groups. Since acetylation was identified in 1960s, more than eight kinds of acylation modifications have been discovered, especially after 2009, because of the quick development of mass spectrometry and biochemistry technologies, as well as powerful algorithm methods. The eight kinds of protein acylations mentioned here can be divided into three groups according to their chemical structures. Acetyl- and crotonyl- are short-chain hydrophobic acyl groups. Myristoyl- and palmitoyl- are long-chain fatty acid hydrophobic acyl groups. β-hydroxybutyryl- and lactyl- belong to the polar acyl groups. Succinyl- and malonyl- belong to the negatively charged acidic acyl groups. The short-chain acylation mainly occurs at lysine residues. Whereas myristoylation often occurs at N-terminal glycine or lysine residues, and palmitoylation usually occurs at cysteine, serine or N-terminal amino acid residues
Donors, writers, erasers and readers of protein acylation
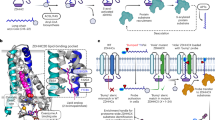
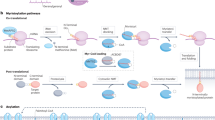
Protein acylation is regulated either in a nonenzymatic or enzymatic manner with the latter one more common. In the enzymatic dependent condition, the acyltransferase—“writer” is responsible to add acyl groups from the “donors” of acyl-CoA, including acetyl-, succinyl-, malonyl-, crotonyl-, β-hydroxybutyryl-, lactyl-, myristoyl-, and palmitoyl-CoA, to the side-chain of lysine (glycine, cysteine, serine or others) residues. The deacylase—“eraser” catalyzes the removal of acyl groups from the aforementioned amino acid residues. The acylation marks are usually read by the specific protein domains, sometimes referred to as “reader” (Table 1).
Donors and the biological function of protein acylation
Acyl-CoA often acts as acyl donor for protein acylation, which are mainly derived from metabolites of glucose, fatty acid, and amino acid. The protein acylation level has a close relationship with cellular acyl-CoA concentration; therefore it is dynamically regulated by metabolic state such as feeding and starvation.14 The differences of acyl-CoA chemical structures mainly contribute to their different influences on the physiochemical properties of the substrate proteins, such as the hydrophobicity, electric charge, steric hindrance and so on. The eight kinds of protein acylation mentioned in this review can be divided into three groups according to their chemical structures: (1) hydrophobic acyl groups, including short-chain acylation of acetyl and crotonyl; long-chain lipid acylation of palmitoyl and myristoyl, which increase protein hydrophobicity and membrane binding ability. (2) Negatively charged acidic acyl groups, including malonyl and succinyl, which change the charge at the K residue from +1 to −1 without disturbing the physiological pH level.15 (3) Polar acyl groups, including β-hydroxybutyryl and lactyl, which contain a hydroxyl group to enable the formation of hydrogen bonds with other proteins.4 We will then describe the source of each kind of donor in detail.
Donors of protein acetylation
As the donor of protein lysine acetylation (Kace), acetyl-CoA is a central metabolite and substrate for anabolic metabolism, which has been studied thoroughly. It can be produced in the mitochondria, cytoplasm or nucleus. Mitochondrial acetyl-CoA is derived from glycolysis, lipid β-oxidation, and the catabolism of branched amino acids (i.e., valine, leucine, and isoleucine). In glycolysis, mitochondrial pyruvate is decarboxylated to form acetyl-CoA, CO2, and nicotinamide adenine dinucleotide hydride (NADH) by the pyruvate dehydrogenase complex (PDC).16 In β-oxidation, the acyl-CoA synthetase protein family catalyzes the CoA- and adenosine triphosphate (ATP)-dependent conversion of cytosolic free fatty acids into acyl-CoA.17 In amino acid metabolism, branched-chain amino acids are first transformed to branched-chain α-ketoacids and then catalyzed into NADH, acetyl-CoA, and other acyl-CoA thioesters.18 Besides the three ubiquitous metabolic circuitries mentioned above, acetyl-CoA also comes from organ-specific pathways. It could derive from β-hydroxybutyrylate in neuron or from ethanol–acetaldehyde–acetate axis in liver cells or brain cells.19,20,21 Glycolysis- or β-oxidation-derived mitochondrial acetyl-CoA represents the major source of cytosolic acetyl-CoA upon transportation. In addition, cytosolic acetyl-CoA can also be derived from glutamine reductive carboxylation, especially when glycolysis is blocked and from acetate in an ATP-dependent manner.22,23 The acetyl-CoA in the nucleus is freely diffused from the cytosol. Besides, two acetyl-CoA-generating enzymes, namely ATP-citrate lyase (ACLY) and acyl-CoA synthetase short-chain family member 2 (ACSS2), are localized in the nucleus and linked to cell growth and proliferation.24,25
Biological function of protein acetylation
All lysine acetyltransferases (KATs) require acetyl-CoA as donor for acetylation reactions. Reciprocally, protein lysine acetylation represents an important mechanism to regulate overall energy metabolisms through either metabolic enzymes or transcription factors. A proteomic analysis of lysine acetylation in rat islets revealed that almost all enzymes in core metabolic pathways related to insulin secretion were acetylated in response to high glucose.26 For example, glucose increased the acetylation of trifunctional enzyme subunit alpha (ECHA, catalyzing the second and third step of long-chain fatty acid β-oxidation) at K644 and K505. Such modifications significantly decreased fatty acid β-oxidation and enhanced the insulin secretion in islet β-cells. As one of the members of the mammalian forkhead box O (FOXO) transcription factor family, FOXO1 is highly expressed in insulin-responsive tissues, including the pancreas, liver, skeletal muscle, and adipose tissue to orchestrate energy homeostasis. FOXO1 acetylation is catalyzed by CREB binding protein (CBP) at K242, K245, and K262. The positive charge of these lysines in FOXO1 contributes to its DNA-binding activity; and acetylation at these residues reduces its binding ability to DNA sequence and attenuates its transcriptional activity.27 FOXO1 acetylation is regulated during the feed-fast cycles.28 In fasting status, hepatic ETS Proto-Oncogene 1 (ETS1) expression is suppressed through MEK-ERK pathway, allowing FOXO1 nuclear trapping and glucogenetic genes transcription. During the feeding period, elevated ETS1 cooperates with CBP to induce FOXO1 acetylation via enhancing their association. Such effect promotes FOXO1 nuclear export and suppresses hepatic gluconeogenesis. Exercise is a good way to boost metabolism. After exercise, widespread protein lysine acetylation could be observed in the skeletal muscle, which is critical for muscle contraction and structure.29
Protein kinases bind ATP and use it to phosphorylate other proteins. Acetylation is known to regulate kinase activity through impairing or enhancing ATP binding. Generally, there is a conserved lysine residue in the ATP-binding pocket of protein kinases, which could be acetylated to affect kinase catalytic activity. Cyclin-dependent kinase 5 (CDK5) is highly expressed in the brain and plays a role in regulating axonal and dendritic growth, neuronal migration, and synapse development. CDK5 acetylation at K33 by general control nonrepressed-protein 5 (GCN5) leads to the loss of its kinase activity via impairing the ATP binding, negatively regulating neurite outgrowth and determining neurite length.30 Notably, acetylation of the conserved lysine residue in ATP-binding pocket is not always impairing the ATP binding. P38 mitogen-activated protein kinase (MAPK) was reported to be reversibly acetylated by p300/CREB binding protein-associated factor (PCAF)/p300 and histone deacetylases 3 (HDAC3) at K53, a lysine site located in its ATP-binding pocket.31 Acetylation of K53 increases the binding affinity of p38 with ATP and enhances its kinase activity. These observations suggest that acetylation at the conserved lysine in the ATP-binding pocket could be a mechanism in controlling kinase activity. However, why lysine acetylation increases the ATP-binding activity in some kinases but decreases it in others still needs further investigation.
Donors of other protein acylation
The research on acyl-CoA of other short-chain acylations including lysine malonylation (Kmal), succinylation (Ksuc), crotonylation (Kcro), β-hydroxybutyrylation (Κbhb) and lactylation (Klac) or long-chain acylation including myristoylation and palmitoylation is not so thorough and enough. Until now, there are mainly three sources of acyl-CoA for these short-chain acylation: (1) short-chain fatty acid (SCFA) or carboxylic acid, including malonate, succinate, crotonate, β-hydroxybutyrate, lactate and α-ketoglutarate (α-KG). Under insufficient glucose energy supply and the fatty acid mobilization conditions, SCFA is transfered into acyl-CoA via ACSS2 to enhance lipid β-oxidation and protein acylation of malonylation, succinylation, crotonylation and β-hydroxybutyrylation. The lactyl-CoA level is upregulated in the condition of anaerobic glycolysis. Besides, succinyl-CoA is mainly derived from α-KG via the α-KG dehydrogenase complex (α-KGDHC) in the tricarboxylic acid (TCA) cycle.32,33,34,35 (2) Amino acids metabolism. Acyl-CoA also derived from amino acid catabolism. Isoleucine, methionine, and valine could be transformed into succinyl-CoA. Lysine, hydroxylysine, and tryptophan could be transformed into crotonyl-CoA.36 (3) Other kinds of acyl-CoA. Acyl-CoA for the short-chain acylation could also be produced by other acyl-CoA via carboxylases. For example, acetyl-CoA could be catalyzed by acetyl-CoA carboxylase (ACCase) into malonyl-CoA.37 Crotonyl-CoA could be transformed from Glutaryl-CoA by Glutaryl-CoA dehydrogenases (GDHs) through dehydrogenation and decarboxylation.38 In the long-chain fatty acid acylation, the myristoyl- and palmitoyl-CoA are often derived from long-chain fatty acid existed in the edible oil such as coconut oil, butter and palm oil.
Biological function of protein succinylation and malonylation
SIRT5 acts as desuccinylase, demalonase and deglutarylase. Sirt5−/− mice were widely used to study the role of succinylation and malonylation. The three kinds of SIRT5-regulated acylations are all connected with metabolism regulation, such as fatty acid oxidation. A systematic profiling of the mammalian succinylome revealed potential impacts of lysine succinylation on enzymes involved in mitochondrial metabolism, the TCA cycle, and fatty acid metabolism.39 Succinate dehydrogenase (SDH) catalyzes the sixth step of TCA cycle to convert succinate into fumarate. SDH succinylation activates its enzymatic activity, suggesting a self-regulatory mechanism of succinate levels in mitochondria.39 Another metabolomics-assisted proteomic study identifies protein lysine succinylation predominantly accumulates in the heart when Sirt5 is deleted, suggesting succinylation in the regulation of heart metabolism and function.40 Here, the succinylation impairs fatty acid oxidation through downregulation of ECHA activity, resulting in lower cardiac ATP production and heart dysfunction during energy-demanding situations such as fasting and exercise. Malonyl-CoA is a tightly regulated metabolic intermediate, which is produced by acetyl-CoA carboxylase and consumed by malonyl-CoA decarboxylase (MCD), fatty acid synthase, and fatty acid elongases. Using MCD−/− cells as a model, increased lysine malonylation was found to show impaired mitochondrial respiration and fatty acid oxidation.41 These studies indicate that succinylation or malonylation of metabolic enzymes function as a crosstalk mechanism between metabolic processes and nutrient change.
Biological function of protein β-hydroxybutyrylation and crotonylation
β-hydroxybutyrate (β-OHB) is the most abundant ketone body. Besides oxidation as an energy substrate, β-OHB is involved in PTMs of histone and nonhistone proteins. Starvation and ketogenic diet stimulate global protein Kbhb in the liver and kidney. Several enzymes of the methionine cycle were β-hydroxybutyrylated in the liver, suggesting that protein β-hydroxybutyrylation may play a role in methionine homeostasis under metabolic stresses, such as prolonged fasting and ketogenic diet.42 Different from the tissue specific distribution of β-hydroxybutyrylation, crotonylation of nonhistone protein is widely distributed in subcellular compartments and affects diversity of protein function, such as gene transcription, DNA damage response, enzymes regulation and metabolic pathways.43 Crotonate, mainly produced by the colon microbiota, is the SCFA precursor of crotonyl-CoA. From this aspect, crotonylation can be considered as a link between the host and gut microbiota.
Biological function of protein palmitoylation and myristoylation
Palmitoylation and myristoylation represent the two most common protein lipid modifications. Myristoylation is an important protein modification in the immune response catalyzed by N-myristoyltransferase (NMT). Thymus is the primary site for T-cell development and has been shown to have high NMT activities. Myristoylation is an essential lipid modification in the thymus during T-cell development.44 In addition, myristoylation is indispensable for the formation of immunological synapse. Myristoylation of Lck and Fyn is necessary for their localization to the immunological synapse, allowing the activation of T-cell receptor (TCR) signaling.45 N-myristoylation is also involved in the control of innate immunity. TRIF-related adaptor molecule (TRAM) is an adaptor molecule exclusively function in the Toll-like receptor 4 (TLR4) pathway. Myristoylation of TRAM targets it to the plasma membrane, where it is essential for the LPS-induced release of inflammatory mediators and cytokines through the TLR4 signaling.46 Similar to myristoylation, palmitoylation is also crucial for protein-membrane docking. For example, Src protein requires both myristoylation and palmitoylation together to form a “dual signal” motif that targets them to membranes.47 In fact, palmitoylation is well-known to be highly prevalent among neuronal proteins and may be relevant to the processes of learning and memory.48,49
Writers of protein acylation
Writers of protein acylation mainly include KAT family, zinc finger aspartate–histidine–histidine–cysteine (DHHC)-type containing (ZDHHC) family and NMT family. KAT family was first regarded as writers of acetylation. Whereas, with the discovery of novel protein acylation types, many members of KAT family were shown to have an expanded repertoire of other short-chain acyltransferase activities. ZDHHC and NMT families are mainly responsible for palmitoylation and myristoylation.
Writers of protein acetylation
Approximately 17 human KATs have been identified of histone acetyltransferases (HATs), which can be divided into five families based on the degree of sequence similarity. The KATs consist of the GCN5-related N-acetyltransferases family (GNAT), which are represented by GCN5 and PCAF; the p300/CBP family, including p300 and CBP; the MYST family, which is represented by MOZ, MOF, Ybf2 (Sas3), Sas2 and TAT interacting protein 60 (Tip60), and monocytic leukemia zinc finger protein; the steroid receptor coactivator (SRC) family, which is represented by SRC-1, 2 and 3; and acetyltransferases, which can not be clearly categorized based on defining features of the first four classes, such as acetyl-CoA acetyltransferase 1 (ACAT1).
Writers of other short-chain protein acylation
P300/CBP is the common writer for almost all short-chain protein acylations, as it has a deep aliphatic pocket within the active site, a critical feature to bind with bulk acyl groups that is not observed in other HATs.50 Members of the GNAT and MYST families have more limited range of acylation activities. For example, Kcro is written by MOF and GCN5, Kmal and Ksuc are written by GCN5 besides p300.51,52,53,54,55 Except for the HATs, Kmal and Ksuc separately have their specific writers of phenolic glucoside malonyl-transferase 1 (PMAT1) and carnitine palmitoyl transferase 1A (CPT1A).56,57 In a study of SIRT5-regulated lysine malonylome using label free quantitative proteomics, researchers found that 56% of mitochondrial Kmal sites overlapped with previously identified Ksuc sites, and 44% of Kmal sites were distinct from both succinylation and acetylation. This is obviously distinct to the fact that succinylation sites overlap with 80% sites of other acylations. They speculated that there might be one acyltransferase utilizing both acetyl-CoA and succinyl-CoA while another acyltransferase possessing high selectivity of malonyl-CoA. It is rational to guess that many specific writers are still waiting to be found behind the “acylation code”.58 Notably, many acylations are regulated both in nonenzymatic and enzymatic manners such as Kace, Ksuc, and Kcro. The nonenzymatic process is mainly controlled by the concentration of acyl-CoA in mitochondria, pH, and protein parameters.59,60
Writers of protein myristoylation and palmitoylation
In contrast to the lysine residue-mediated post-translational modifications mentioned above, protein myristoylation mainly occurs on the N-terminal glycine or lysine residues via a stable amidic linkage catalyzed by NMT1 and NMT2. Protein palmitoylation is mainly categorized into S-palmitoylation (cysteine residues, Spalm), O-palmitoylation (serine residues), and N-palmitoylation (N-terminus).61 Spalm is mediated by the palmitoyl-acyltransferases (PAT) of ZDHHCs, which contain 23 distinct members in mammals. Mechanistically, the cysteine residue of the DHHC domain reacts with palmitoyl-CoA, forming an acyl intermediate, and then transfers it directly to the substrate protein.
Erasers of protein acylation
Erasers for protein acetylation
HDACs are enzymes that catalyze the removal of acetyl functional groups from the lysine residues of both histone and nonhistone proteins. HDACs can be divided into two categories, Zn2+-dependent and NAD+-dependent HDACs. Zn2+-dependent HDACs include class I (HDAC1, 2, 3, and 8), II (IIa: HDAC4, 5, 7, and 9; IIb: HDAC6 and 10) and IV (HDAC11) subgroups, while NAD+-dependent enzymes are class III HDACs (SIRT1-7).62,63,64
Erasers for other short-chain protein acylation
The short-chain protein acylation share the deacylases of HDAC I, II, and III proteins due to their similar chemical structures of amide bond. The removal of protein Kmal and Ksuc, the negatively charged acidic acyl groups, is mainly dependent on SIRT5, which is closely related to some diseases of cancer and neurodegenerative diseases.65,66,67 According to the crystal binding structure of succinyl-Lys peptide and SIRT5, researchers found that tyrosine 102 and arginine 105 are the special residues of SIRT5, in the deep end of substrate-binding pocket, which forms hydrogen bonds and ionic bonds with the carboxyl group of the succinyl lysine substrate. This might also suit for its binding with malonyl lysine substrate. Besides SIRT5, SIRT7 was also reported as a desuccinylase, which has close relationship with DNA damage.68 Whether SIRT7 could regulate the other two negatively charged acidic acyl groups is still unknown. The removal of S-form and R-form β-hydroxybutyryl, the polar acyl groups, is mediated by SIRT3 and HDAC1/HDAC2, respectively.69,70 The distinct backbone sensitivity of NAD+-dependent (SIRT3) and Zn+-dependent (HDAC1/HDAC2) subfamilies decides their chiral selectivity, which still needs in-depth study. Similar with Kbhb, HDAC1–3 and SIRT1–3 have been identified as delactylases in vitro. Besides, the de-L-lactylase activity of HDACs 1 and 3 have been verified in cells.71 The erasers of the hydrophobic crotonyl group are mainly class III HDACs, including SIRT1, 2, and 3.72
Erasers for protein myristoylation and palmitoylation
Depalmitoylation is carried out by serine hydrolases, including acyl-protein thioesterase (APT), palmitoyl protein thioesterase (PPT), and a family of mammalian α/β hydrolase domain-containing proteins (ABHDs). Erasers of lysine or N-terminal glycine myristoylation are totally different. The removal of lysine myristoylation is mainly mediated by SIRT6, which has a large hydrophobic pocket that makes it a perfect eraser for long-chain fatty acyl groups.73 N-terminal glycine myristoylation has long been regarded as an irreversible protein lipidation. However, in a recent study, researchers identified a new demyristoylase for human ARF1—invasion plasmid antigen J (IpaJ), a previously uncharacterized Shigella flexneri type III effector protein with cysteine protease activity.74
Readers of protein acylation
Proteins containing either of the following five domains have been characterized as histone and nonhistone acylation readers and the bank is still being expanded.4,75,76,77 The first category are proteins with bromodomains (BDs), which include bromodomain and extra-terminal (BET) or non-BET family of proteins. The second are proteins with double PHD finger (DPF), including MOZ, MOF, and DPF2. The third are the YEATS family proteins, including AF9, YEATS2, GAS41, TAF14, Sas5, Yaf9, and ENL. The last two are proteins with double PH domain (such as Rtt106) and ZZ-type zinc finger domain (such as p300). Besides, many “writers” have dual identities as “readers” such as CBP (bromodomain), p300 (ZZ domain), MOF (DPF domain), and MORZ (DPF domain).
Studies on protein acylation readers are mostly focused on histone Kace, Kpro, Kcro, Ksuc, Kbhb, and nonhistone Kace. Few reports are on the readers of histone or nonhistone protein long-chain fatty acid lipidation, studies are still in need to explore whether there are unknown readers to bind such bulk acyl groups. One reader can recognize different types of protein acylation with its unique priority. For example, many readers of the YEATS type bind to K acylation through an aromatic cage, with the highest affinity towards crotonylation followed by the other acyl marks approximately in order of the length of the fatty acid chain.78,79,80 The readers usually translate the protein acylation marks to the signal in regulating protein transcriptional activity, DNA-binding ability, or degradation speed.
Among the readers, only the type of bromodomains (BRD4, BRD3, and PBRM1) have been reported to recognize acetylated nonhistone proteins, which are all transcription factors. BRD4 is a member of the BET family with two BDs (BD1 and BD2) reading acetylated RelA, ERG, Twist, and Snail. Both BD1 and BD2 domains of BRD4 could recognize and interact with RelA, recruiting CDK9 to phosphorylate the C-terminal domain of RNA polymerase II and facilitating the transcription of NF-κB-dependent genes.81,82,83 Similar with the working model of BRD4 on RelA, BRD4 binds with the ERG acetylated 96KGGK99 motif and the Twist diacetylated “histone H4-mimic” GK-X-GK motif to upregulate their transcriptional activity.84 Different from all the mechanisms mentioned above, BRD4 recognizes CBP-acetylated Snail (K146 and K187) to enhance its protein stability.85 BRD3 is another member of BET family with BD1 and BD2 domains. It has been reported to promote erythroid maturation through “reading” the acetylated GATA1 with its BD1 domain and promoting its stable association with chromatin.86 In summary, BRD4 and BRD3 use different BD domains (BD1, BD2, or both) to recognize the acetylated nonhistone proteins and regulate either their transcriptional activity or protein degradation, the selectivity of the recognition domains and the working mode is still being elucidated in the future. PBRM1 is the second most highly mutated tumor suppressor gene in kidney cancer with four BD domains. Recent studies found that PBRM1 reads acetylated K382 of p53 through its BD4 domain to promote the interaction of p53 with the promoter of its target gene p21.87 Whether there are any other readers of the bromodomain type or of other types that are responsible for the recognition of nonhistone protein acetylation and other kinds of acylation still needs to be studied.
Protein acylation in human diseases
Acetylation and its role in human diseases
Acetylation is a metabolic and chemical process, during which the acetyl group is attached to the protein/peptide or messenger RNA.2,60 In protein acetylation, the acetyl groups bind covalently to the lysine, serine or threonine residues of amino acids either in a nonenzymatic manner, especially in alkaline environments such as the mitochondrial matrix, or in an enzymatic manner.88,89,90,91
Protein acetylation in tumor
Protein acetylation and deacetylation mediated by KATs and HDACs can occur in either tumor cells or immune cells and eventually change the intrinsic tumor features or immune cell phenotypes. Hereafter, we summarize and discuss the effects of nonhistone protein acetylation or deacetylation in tumor development and progression that have been reported in the last 10 years.
Protein acetylation in the metabolic adaptation of tumors
Protein acetylation regulates tumor progression by regulating metabolic enzymes, affecting their enzymatic activity or stability (Fig. 2a). Vigorous glycolysis in tumors is closely related to acetylation. Phosphoglycerate kinase 1 (PGK1) and PGK2 are the only enzymes that produce ATP in the glycolysis pathway, in which PGK1 is overexpressed in liver cancer.92,93 Researchers found that PCAF-mediated K323 acetylation of PGK1 enhances its enzymatic activity and glucose uptake, elevating liver cancer cell proliferation, and tumorigenesis.94 Enolase 2 (ENO2) is another key glycolytic enzyme in the metabolic process of glycolysis and is overexpressed in prostate cancer, small-cell lung cancer, metastatic neuroblastoma, and leukemia.95,96 HDAC3-mediated deacetylation of ENO2 at K394 leads to its activation and enhancement of glycolysis, which finally results in the metastasis of pancreatic ductal adenocarcinoma (PDAC).97
Protein acetylation in shaping tumor metabolism and oncogenic signaling. Protein acetylation usually influence tumor progression via regulating metabolic enzymes and oncoproteins. a Protein acetylation in the regulation of tumor metabolism. Metabolic enzymes responsible for tumorigenesis and proliferation are regulated by acetylation. PGK1 is acetylated by PCAF at K323 to promote glucose uptake. GNPAT is acetylated by ACAT1 at K128 to inhibit FASN degradation and enhance lipid synthesis. SIRT2 degradation leads to succinate production and H3K4me3 activation. The above effects either provide energy source for tumor proliferation or activate tumor-specific gene transcription. Besides, protein acetylation can also occur on metabolic enzymes responsible for tumor metastasis. ENO2 is deacetylated by HDAC3 at K394 to increase its activity and glycolysis. IDH1 involved in glutamine metabolism is deacetylated at K224 to inhibit its enzymatic activity and HIF1α-SRC transcription axis. Enhanced glycolysis and HIF1α-SRC transcription axis is closely connected with tumor metastasis. b Acetylation of oncogenic signaling proteins relates to tumorigenesis, proliferation and metastasis. TRIB3 promotes KAT5-mediated SMAD3 acetylation at K333 to promote the transcriptional activity of SMAD3, which positively regulates transcription of the downstream TRIB3 and results in autophagy blockade. SIRT2 inhibits SMC1A acetylation at K579 to induce proper mitosis. SMAD3 recruits p300 to acetylate KLF5 at K369 and promote the expression of its target gene—CXCR4 and EMT. BRD4 recognizes CBP-acetylated Snail (K146 and K187) to enhance its protein stability and promote EMT. ACC1 is phosphorylated and inactivated by leptin or TGF-β signaling, resulting in increased acetyl-CoA and SMAD2 acetylation, which finally upregulates SMAD2 transcriptional activity and EMT
Acetylation of enzymes in lipid metabolism have also been found to promote cancer development. Glyceronephosphate O-acyltransferase (GNPAT) is critical for the synthesis of fatty acids, which are dysregulated in hepatocellular carcinoma (HCC).98 The acetyltransferase ACAT1 is upregulated in response to extra palmitic acid (PA) and acetylates GNPAT at K128, which represses tripartite motif-containing 21 (TRIM21)-mediated ubiquitination and degradation of GNPAT. The accumulated GNPAT then represses the degradation of fatty acid synthase (FASN) mediated by TRIM21 and promotes fatty acid synthesis and hepatocarcinogenesis.99
Protein acetylation of the key enzymes in glutamine metabolism or in the TCA cycle is another good example in tumor. Isocitrate dehydrogenase 1 (IDH1) is involved in glutamine metabolism, which is located in the cytoplasm and converts isocitrate to α-KG. Researchers found that wild-type IDH1 is hyperacetylated at K224 in colorectal cancer (CRC), promoting CRC progression and liver metastasis. SIRT2 could deacetylate IDH1 at K224 and exhibit a tumor suppression function in a colon cancer cell model by inhibiting IDH1 enzymatic activity and the hypoxia-inducible factor 1α (HIF1α)-SRC transcription axis. The above examples indicate that the acetylation of metabolic enzymes play complicated regulatory role of enzymatic activity. Whether acetylation enhances or inhibits the enzymatic activity may depend on the spatial location of the acetylation site relative to the catalytic domain or degradation-related ubiquitination site.
Protein acetylation in oncogenic or tumor-suppressive signaling
In addition to metabolic enzymes, numerous oncogenic or tumor-suppressive signaling pathways are regulated by acetylation, involving in tumorigenesis, metastasis or drug resistance (Fig. 2b).
The pseudokinase tribbles homolog 3 (TRIB3) has been demonstrated to promote tumorigenesis in lung cancer, liver cancer, breast cancer, colorectal cancer and leukemia.100,101,102,103,104,105,106,107 Recently, our group elucidated that TRIB3 acts as an adaptor to recruit KAT5 to SMAD3, inducing phosphorylation-dependent SMAD3 K333 acetylation. This modification sustains SMAD3 transcriptional activity and subsequently enhances TRIB3 transcription, forming a positive feedback regulation loop. Metformin could inhibits KAT5-mediated SMAD3 K333 acetylation and TRIB3 expression, inhibiting the progression of melanoma.108 Protein acetylation also regulates mitotic catastrophe in cancer. Mitotic catastrophe can be defined as an oncosuppressive mechanism that eliminates mitosis-incompetent cells.109,110 SIRT2 is upregulated in early-stage carcinomas and deacetylates the structural maintenance of chromosome protein 1 (SMC1A), which then promotes its phosphorylation to properly drive mitosis. This permits tumor cells to escape mitotic catastrophe, thus allowing early precursor lesions to overcome oncogenic stress.111
Epithelial–mesenchymal transition (EMT) is a complex developmental program that enables cancer cells to acquire a more aggressive phenotype for metastasis and therapy resistance. Extensive crosstalk occurs between metabolism and EMT to coordinate tumor progression. The lipogenic enzyme acetyl-CoA carboxylase 1 (ACC1) was recently demonstrated to suppress breast cancer migration and invasion in a manner that was independent of fatty acid synthesis but was dependent on acetyl-CoA. Mechanistically, ACC1 is phosphorylated and inactivated by leptin or transforming growth factor-β (TGF-β) signaling, which is highly expressed in obesity. This results in increased acetyl-CoA and SMAD2 acetylation, which finally upregulates its transcriptional activity and promotes EMT programs in breast tumor cells.112 DOT1L is a histone methyltransferase that regulates various genes involved in cancer onset and metastasis.113,114 CBP promotes DOT1L acetylation at K358 and enhances its stability by preventing the binding of RNF8 and DOT1L. The stabilized DOT1L catalyzes the H3K79 methylation of genes involved in EMT, including Snail and ZEB1, thus promoting CRC metastasis.115 Krüppel-like Factor 5 (KLF5) is a key transcriptional factor in regulating cell proliferation, apoptosis, tumor cell stemness traits and EMT.116,117 Acetylation of KLF5 has been reported to play opposite roles in the progression of prostate cancer and breast cancer via regulating its transcriptional activity or protein stability. In prostate cancer (PCa), bone-borne TGF-β was found to promote the acetylation of KLF5, leading to osteoclastogenesis and chemoresistant bone metastatic formation. Mechanistically, SMAD3 recruits p300 to acetylate KLF5 at K369 and enhance its transcriptional activity, thus promoting the expression of its target gene—CXCR4. The increased CXCR4 then promotes IL-11 secretion and stimulates metastasis-associated SHH/IL-6 paracrine signaling.118 While, acetylation of KLF5 at the same lysine residue inhibit tumor progression in basal-like breast cancer (BLBC). Kong et al. found that HDAC inhibitors increase KLF5 acetylation at K369 to interrupt its association with BRCA1-associated protein 1 (BAP1) (a deubiquitinase), promoting its ubiquitination and degradation. This will decrease cell viability in BLBC cell lines.119 It is possible that there is a different secondary signal involved in the two types of tumors, leading to contradictory tumor biological outcomes, which deserves further investigation.
The most deeply studied tumor suppressor under acetylation regulation is p53. In response to cellular stresses, p53 transcriptionally participates in the modulation of multiple biological processes, including proliferation, cell cycle arrest, programmed cell death (apoptosis), cellular senescence, DNA repair, autophagy, oxidative response, and metabolic regulation.120 The acetylation of different lysine residues of p53 plays opposite roles in tumor progression.121 In response to DNA damage, p300/CBP mediates C-terminal (K370, K372, K373, K381, K382, and K386) and K101 acetylation to suppress p53-dependent apoptosis and ferroptosis.122,123 In addition, PCAF-mediated p53 K320 acetylation also promotes cell survival and inhibits apoptosis by selectively inducing the expression of antiapoptotic genes and repressing proapoptotic genes.65 In contrast, p300/CBP and MYST family (TIP60, MOF, and MOZ)-mediated acetylation of p53 at K120 and K164 inhibit tumor progression by promoting apoptosis. Cancer cell-intrinsic programmed cell death 1 (PD-1) has been suggested to suppress lung cancer progression. In a recent study, researchers demonstrated that PD-1 is a target gene of p53, and acetylation of p53 at K120 and K164 helps to promote PD-1 transcription and suppress lung cancer development in an immunity-independent manner.124
Tumor cell protein acetylation in shaping antitumor immune responses
Recently, protein acetylation in tumor cells or immune cells has been reported to shape the immunosuppressive tumor microenvironment by regulating immune cell exhaustion, activation, and infiltration.
Programmed cell death 1 ligand 1 (PD-L1), which is expressed on cancer cells, binds to the receptor PD-1 on T cells to prevent their proliferation and reduce the anti-tumor immune response, resulting in T-cell exhaustion.125,126,127 Researchers found that the acetylation of PD-L1 or related transcription factors could regulate its subcellular localization. Through interacting with vimentin and importin α1, PD-L1 shuttles from the plasma membrane into the nucleus, in which the deacetylation of PD-L1 by HDAC2 at K263 is a precondition.128 Nuclear PD-L1 then directly binds with DNA to regulate the transcription of multiple immune response-related genes and several immune checkpoints. Blocking PD-L1 nuclear translocation via HDAC2 inhibition enhances the PD-1 blockade therapy. Protein acetylation also affects PD-L1 transcription levels. Myocyte enhancer factor 2D (MEF2D) is a transcription factor that is overexpressed in HCC and is associated with poor survival of HCC patients.129 Researchers found that when HCC cells are exposed to interferon gamma (IFN-γ), p300 acetylates MEF2D and promotes its binding with the PD-L1 promoter, leading to increased PD-L1 expression. Strategies to manipulate this pathway might increase the efficacy of immune therapies for HCC (Fig. 3a).130,131
Protein acylation in shaping tumor immune microenvironment. Protein acylation helps to shape immunosuppressive tumor microenvironment via regulating immune cell activation, infiltration and antigen presentation. a Protein acylation in immune braking or activation. HDAC2 inhibits PD-L1 acetylation to increase its nuclear localization and immune checkpoints activation. P300 mediates MEF2D acetylation to promote PD-L1 transcription. ZDHHC3 and ZDHHC9 mediate PD-L1 palmitoylation to inhibit its lysosomal degradation. The three events will induce T cells exhaustion. Rae-1 is acetylated by PCAF and GCN5 to enhance its stability and activate NK/T cells killing ability. b Protein acylation in immune infiltration. P300-mediated TRIB3 acetylation inhibits T cells infiltration through inhibiting CXCL10 transcription. SIRT1-mediated p53 deacetylation promotes TAM infiltration through secreting CXCL12. KAT6A-mediated SMAD3 acetylation results in its transactivation and the transcription of cytokines, including IL-6/IL-12/TNF-α and promotes MDSC infiltration. c Protein acylation in antigen presentation. OPTN interacts with AP3D1 to hinder its recognition of IFNGR1, thereby maintaining IFNGR1 stability and the integrity of downstream MHC-I signaling, promoting antigen presentation to T cells
In contrast to PD-L1, the natural killer group 2 member D (NKG2D) ligand Rae-1 binds with NKG2D in T cells or NK cells to activate their tumor-killing activity.132 Shedding, loss of expression, or internalization of ribonucleic acid export 1 (Rae-1) from the tumor cell surface leads to immune evasion, which is associated with poor prognosis in patients with cancer.133,134,135 Researchers found that GCN5 and PCAF acetylate Rae-1 at K80 and K87 to enhance its stability and protect Rae-1 from shedding to activate the immune surveillance of NK cells and CD8+ T cells. This observation may shed light on new targets for NKG2D immunotherapy in cancer treatment (Fig. 3a).136
Protein acetylation in tumor cells also influences immune cell infiltration by regulating the secretion of chemokines. SIRT1 was demonstrated to promote CXCL12 expression by inhibiting the acetylation of p53 in colorectal cancer. This induces tumor-associated macrophage (TAM) migration through the CXCL12/CXCR4 pathway. The recruited TAMs further inhibit the proliferation and activity of CD8+ T cells, resulting in CRC progression.137 In some cases, the acetylation of oncoproteins could regulate both cancer stemness features and immune cell recruitment at the same time. Our recent study revealed that p300 induces TRIB3-K240 acetylation under high-glucose conditions. This modification attenuates the association of TRIB3 with the E3 ligase SIAH1 and inhibits its ubiquitination and degradation.138 On one hand, aberrant highly expressed TRIB3 enhances the stemness of cancer cell by activating the Wnt/β-catenin signaling pathway.102 On the other hand, TRIB3 represses the STAT1-CXCL10 axis and reduces CD8+ T-cell infiltration, resulting in the immune evasion of CRCs. This work highlighted a potential therapeutic target for treating immunologically “cold” CRCs.101 The acetylation of SMAD3 is another good example. SMAD3 often acts as a mediator of TGF-β superfamily that modulates signaling and has been implicated as a driving event in cancer metastasis.81 The acetylation of SMAD3 at K20 and K117 by KAT6A promotes its association with oncogenic chromatin modifier TRIM24 and disrupts its interaction with the tumor suppressor TRIM33, enhancing the transcription of immune response-related cytokines. This enhances myeloid-derived suppressor cell (MDSC) recruitment and breast cancer stem-like cell stemness, which promote triple-negative breast cancer (TNBC) metastasis (Fig. 3b).139
Immune cell protein acetylation in shaping of antitumor immune responses
Protein acetylation in immune cells also regulates tumor-killing activity. STAT6 acetylation in TAMs affects their polarization and tumor-killing ability. STAT6 is known to drive macrophage M2 polarization. The K383 of STAT6 is acetylated by the acetyltransferase CBP during macrophage activation to suppress macrophage M2 polarization. Mechanistically, TRIM24, a CBP-associated E3 ligase, promotes STAT6 acetylation by catalyzing CBP ubiquitination at K119 to facilitate the recruitment of CBP to STAT6. In contrast, STAT6 mediates the suppression of TRIM24 expression in M2 macrophages, contributing to the induction of an immunosuppressive tumor niche.59,140
Protein acetylation in Alzheimer’s disease
Alzheimer’s disease (AD) is the most common neurodegenerative disease, which is pathologically caused by neurofibrillary tangles (NFTs) and extracellular accumulation of beta-amyloid (Aβ). In AD, highly phosphorylation and aggregation of tau protein contribute to the formation of NFTs and to mediate Aβ toxicity. Therefore, reducing tau is a prospective strategy for AD therapy. Tau is majorly degraded by macroautophagy and chaperone-mediated autophagy (CMA) to avoid its excessive accumulation in AD.141 Acetylation of soluble tau is an early pathological event in neurodegeneration. Numerous studies have reported the complex relationship between tau acetylation and its protein stability. Acetylation of tau at K174 or K274 impedes CMA, which alters tau homeostasis and contributes to aggravate disease progression.142 Another group demonstrated that the loss-of-function mutation in the TSC Complex Subunit 1 (TSC1) gene results in upregulated p300 and damaged SIRT1 enzymatic activity, leading to tau acetylation and preventing tau clearance via CMA.143 HDAC6 was reported to deacetylate and inhibit hyperphosphorylation of tau, which alleviates neurodegeneration and cognitive decline.144 While another group has discovered the opposite relationship between tau acetylation and its protein stability. Choil et al. found that acetylation of tau at K274, K290, K321, and K353 recruits chaperone proteins, including Hsp40, Hsp70, and Hsp110, which facilitates novel tau E3 ligases binding and tau degradation.145 The contradictory phenomena might due to the different acetylated lysine residues in tau, which indicate the complicated regulatory network of tau PTMs.
Protein acetylation in hepatic steatosis
The liver is the metabolic hub of glucose, fatty acid, and amino acid, which is largely affected by the metabolic enzymes and associated PTMs. Protein acetylation has been linked with both alcoholic (AFLD) and non-alcoholic fatty liver disease (NAFLD) through regulating transcription factors or metabolic enzymes. SIRT2 has been demonstrated to play opposite roles in AFLD and NAFLD via regulating different transcription factors. SIRT2 mediates CCAAT/enhancer-binding protein beta (C/EBPβ) deacetylation at K102 and K211 to inhibit its ubiquitination and degradation.146 The accumulated C/EBPβ promotes ethanol-induced liver injury via regulating the expression of target genes involved in adipogenesis, gluconeogenic pathway, liver regeneration, and so on. While, in the NAFLD, SIRT2 prevents liver steatosis and metabolic disorders by deacetylating and stabilizing hepatocyte nuclear factor 4α (HNF4α) at K458.147 In both of the two studies, Kace works on the transcription factors via regulating their protein stability but not transcriptional activity, providing a novel regulatory mechanism of acetylation on transcription factors. Besides, it’s because of the different functions of the substrates that make similar stability regulation by SIRT2-mediated deacetylation with opposite effects on disease progression. Except for the transcriptional factors, the acetylation of key enzymes in glucose metabolism—lactate dehydrogenase-B (LDHB) also regulates NAFLD. PCAF-mediated acetylation of LDHB at K82 was found to significantly decrease its enzymatic activity and impair hepatic lactate clearance, resulting in lactate accumulation, which exacerbates lipid deposition and inflammatory responses by activating histone hyperacetylation in high-fat diet (HFD)-induced NAFLD.148 All of the evidence suggests the importance and complex of Kace in regulating metabolic diseases and provides potential therapeutic targets from the view of protein PTMs.
Protein acetylation in immune and infectious disease
Protein acetylation has close relationship with immune response in chronic inflammation and virus infection. In the aging-associated chronic inflammation, the acetylation of the inflammasome NLRP3 in macrophages is critical to its assembly and activation, leading to the production of inflammatory cytokines IL-1β and IL-18.149 In the innate immune response to DNA virus, the DNA sensor cGMP-AMP synthase (cGAS) senses cytosolic microbial or self DNA to initiate a MITA/STING-dependent innate immune response. Acetylation of cGAS by KAT5 in its N-terminal domain promotes its DNA-binding ability, increasing the transcription of downstream antiviral genes.150 In addition to the DNA virus infection, acetylation of OTU deubiquitinase 3 (OTUD3) has been verified to be critical to the RNA virus infection.151 Since 2019, SARS-CoV-2 has caused an ongoing pandemic of coronavirus disease 2019 (COVID-19) worldwide.152 Protein acetylation has also been studied in this severe acute respiratory syndrome. Acetylated K676 of TGF-β-induced protein (TGFBIp) was consistently elevated in the blood of patients with SARS-CoV-2 pneumonia, especially in patients of the intensive care unit (ICU) compared to non-ICU patients, suggesting it as a severity diagnostic biomarker.153 Although the mechanisms are still unclear, it can be predicted that protein Kace might be a potential prognostic or therapeutic target of the immune or infectious diseases.
Succinylation and its role in human diseases
Succinylation has mainly occurred in the mitochondria which was identified and verified by Zhang et al.6 in 2011 by mass spectrometry and protein sequence alignment in vivo.85,154 Different from acetylation, Ksuc causes greater changes on lysine residues in charge (from +1 to −1) and structure, which is likely to have a significant impact on the substrate proteins. In detail, the positively charged chains of lysine residues in physiological pH plays critical roles in protein folding and the formation of noncovalent interactions such as the leucine zipper, as well as in general acid-base catalyzed enzymatic reactions in which proton transfer is required.155 Therefore, Ksuc is likely to lead to significant changes in PPI, enzymatic or transcriptional activities. Studies have discovered that most of the succinylated proteins are involved in the regulation of energy metabolism and translation, and nearly every enzyme of the TCA cycle is succinylated, implying a possible role for Ksuc in energy metabolism.156 Nonhistone protein Ksuc has been studied in tumor, COVID-19, metabolic diseases, neurological disease, cardiovascular disease, immune system diseases, and mitochondrial diseases.157
Protein succinylation in tumor
Nonhistone Ksuc mainly regulate tumor development through influencing metabolic reprogramming, oncogenic signaling pathways and suppressive immune microenvironment.
Protein succinylation in the regulation of tumor anaerobic glycolysis and serine metabolism
LDHA mediates the last step in the anaerobic oxidation of glucose and catalyzes the formation of lactate. Protein succinylation mediated by CPT1A has been demonstrated to promote the proliferation and invasion of gastric cancer (GC) by regulating LDHA succinylation. Mechanistically, CPT1A succinylates LDHA at K222, which thereby reduces the binding of K63-ubiquitinated LDHA with SQSTM1 and inhibits its autophagic degradation. Overexpression of a succinylation-mimic mutant of LDHA could promote tumor cell proliferation, migration and invasion (Fig. 4a).158 SIRT5 is both a deacetylase and desuccinylase. In CRC and osteosarcoma, SIRT5 desuccinylates mitochondrial serine hydroxymethyltransferase (SHMT2) at K280 to increase its enzymatic activity, driving serine catabolism and promoting tumor proliferation.159 These results suggested that protein succinylation is used by cancer cells to adapt to metabolic status for rapid growth.
Protein succinylation and malonylation on metabolic enzymes or kinases in tumor, inflammatory, cardiovascular and metabolic diseases. The negatively charged acidic acyl groups including malonyl group derived from acetyl-CoA or malate, and succinyl group derived from α-KG or amino acids take part in the PTMs of metabolic enzymes in numerous kinds of diseases. a, e Protein Ksuc and Kmal in tumor. CPT1A mediated LDHA succinylation at K222 to inhibit its autophagic degradation via p62 to accelerate gastric cancer (a). Depletion or inhibition of FASN enhances malonyl-CoA level and promotes mTOR malonylation at K1218 to downregulate its kinase activity and the subsequent phosphorylation of p70S6K/4EBP1, promoting endothelial cells proliferation and tumor angiogenesis (e). b, f Protein succinylation and malonylation in metabolic enzymes play critical roles in LPS-induced inflammation of macrophages. LPS inhibits SIRT5 mediated desuccinylation of PKM2 at K311 to inhibit its kinase activity and increase its nuclear translocation by promoting PKM2 tetramer-to-dimer transition. The nucleus PKM2 interacts with HIF1α to promote IL-1β transcription and inflammation (b). LPS stimulation enhances malonyl-CoA level and promotes GAPDH malonylation at K213 in macrophages, leading to its increased enzymatic activity and dissociation from TNFα mRNA, promoting TNFα expression and inflammation (f). c, g Protein succinylation and malonylation in metabolic enzymes play critical roles in cardiovascular disease. Knockout of SIRT5 results in increased Ksuc in ECHA at K315, inhibiting its enzymatic activity and ATP production and promoting hypertrophic cardiomyopathy (c). IDH2 malonylation decreases its enzymatic activity and promotes cardiomyopathy (g). d, h Protein succinylation and malonylation in metabolic enzymes play critical roles in metabolic disease. HDAC1 inhibits SREBP1 succinylation and increases its protein stability, promoting hepatic steatosis (d). SIRT5 mediates several key metabolic enzymes malonylation to promote glycolysis and FAO, inhibiting hepatic steatosis (h)
Protein succinylation and desuccinylation in the regulation of oncogenic signaling
S100 proteins are a family of calcium‐binding cytosolic proteins that possess a wide range of intracellular and extracellular functions and play pivotal roles in tumor migration and invasion. CPT1A-induced S100A10 succinylation at K47 suppresses its ubiquitylation and subsequent proteasomal degradation. Succinylated S100A10 is highly expressed in GC and promotes GC invasion.160 Notably, protein succinylation also demonstrates tumor-suppressive effect in the context of cancer types. Using proteomic technologies, lower lysine succinylation was found in esophageal squamous cell carcinoma (ESCC) than in nonmalignant controls. The decreased succinylation may be attributed to the decreased succinyl-CoA, leading to abnormal metabolism in ESCC. In addition, functional assays through either chemical or genetic approaches demonstrated that hyposuccinylation elevates the migratory ability of ESCC cells both in vitro and in vivo. Using tumor tissue samples from ESCC patients, the authors confirmed that the degree of lysine succinylation was higher in adjacent nonmalignant esophageal epithelium than in cancer tissues. These observations highlight the complicated effect of succinylation modification in cancer biology.161
Protein succinylation in shaping the immunosuppressive tumor microenvironment
Succinylation regulators have an intense relationship with immune cell phenotypes, especially regulatory T cells (Tregs). To systematically study the role of succinylation regulators in tumors, Lu et al. performed a comprehensive pan-cancer analysis on four well-known succinylation regulators (CPT1A, KAT2A, SIRT5, and SIRT7). They found that KAT2A was prominently upregulated in all types of tumors compared to the corresponding normal tissues. SIRT7 was significantly upregulated in nine of ten tumor types and downregulated in colon adenocarcinoma. The expression of CPT1A and SIRT5 showed high heterogeneity in different tumors, which is consistent with the previously reported studies. The researchers performed further analysis on clear cell renal cell carcinoma (ccRCC) since all four regulators are associated with the overall survival of patients with ccRCC. Using analytical bioinformatics, they found that SIRT7-high, SIRT5-low or CPT1A-low might contribute to the infiltration of forkhead box P3+ (FOXP3+) Tregs, which induce an immunosuppressive microenvironment. As SIRT5 and SIRT7 are both regulators of deacetylation and desuccinylation, the regulation of Treg cell infiltration might be related to the changes in these two post-translational modifications.162
Protein succinylation in inflammatory and infectious disease
The internal inflammatory reaction is crucial in killing invaders and avoiding harmful infection. However, if the inflammatory response is out of control or persists for a long time, it will cause diseases such as rheumatoid arthritis, inflammatory bowel diseases and potentially fatal sepsis. Lipopolysaccharide (LPS) strongly increases the intermediate succinate level in TCA cycle of macrophages, which stabilizes HIF1α to induce IL-1β expression, establishing the relationship of succinate and Ksuc with inflammation.163 Mechanistically, LPS-activated macrophages undergo a metabolic shift from dependence on mitochondria-produced ATP to reliance on aerobic glycolysis, where pyruvate kinase M2 (PKM2) is a critical determinant.164,165 They found that LPS results in a reduced desuccinylation activity of SIRT5. This will in turn increase PKM2 Ksuc at K311 to inhibit its kinase activity and increase its nuclear translocation by promoting PKM2 tetramer-to-dimer transition, which leads to the interaction of PKM2 with HIF1α and promotes IL-1β transcription and DSS-induced colitis in mice.166 The studies above suggest a key role of protein PTM in linking metabolic reprogramming and inflammation (Fig. 4b).
Novel antiviral therapies against SARS-CoV-2 and the emerging variants are still in urgent need. Viruses can’t replicate by themselves, hence they have evolved to alter the host cellular pathways and protein PTMs to meet their requirement for replication. After SARS-CoV-2 infection, dramatic upregulation of protein Ksuc in host (Caco-2 cells) was observed by using a quantitative mass spectrometry-based succinylproteomic strategy.167 Most of the hypersuccinylated proteins are enzymes in the TCA cycle, including citrate synthase (CS), aconitase 2 (ACO2), dihydrolipoamide S-succinyltransferase (DLST), succinate-CoA ligase GDP/ADP-forming subunit alpha (SUCLG1), succinate-CoA ligase ADP-forming subunit beta (SUCLA2), succinate dehydrogenase complex flavoprotein subunit A (SDHA), fumarate hydratase (FH), oxoglutarate dehydrogenase (OGDH), and malate dehydrogenase 2 (MDH2), leading to the inhibition of cellular metabolic pathways. Besides, viral nonstructure protein NSP14 is capable to participate in succinylation through interacting with host SIRT5. Indeed, SIRT5 activators and CPT1A inhibitors have been confirmed with potential antivirus effects. This study first provides evidence for the close relationship between metabolism and virus infection from the view of PTMs, which opens a new window for exploring anti-COVID-19 strategy in the future.
Protein succinylation in cardiovascular disease
Heart is the organ with huge energy supply need, which depends on healthy TCA cycle and oxidative phosphorylation. Researchers found that heart has the highest concentration of succinyl-CoA which has close relationship with TCA cycle. Therefore, they began to explore the function and molecular mechanism of Ksuc in ischemic cardiomyopathy or hypertrophic cardiomyopathy. In one study, Sadhukhan et al. found that Sirt5−/− mice develops hypertrophic cardiomyopathy with hyper protein succinylation in the heart. ECHA, a protein involved in fatty acid oxidation, was identified with the most succinylation sites (at 28 lysine residues). In detail, depletion of SIRT5 enhances ECHA Ksuc at K315, which inhibits ECHA enzymatic activity, leading to defective fatty acid metabolism, decreased ATP production, and hypertrophic cardiomyopathy (Fig. 4c).40 In another study, differential protein expression and succinylated lysine residues in myofibrils from failing ischemic cardiomyopathy hearts and non-failing hearts in patients were evaluated. Increased succinyl-CoA synthetase activity and succinyl-CoA turnover were found in ischemic heart failure samples, which results in significant decrease in protein Ksuc.168 From the evidence, one can conclude that protein hypersuccinylaiton promotes hypertrophic cardiomyopathy, while there is a decrease of protein succinylation in the ischemic failure heart. The seemingly contradictory results might because: (1) Heart failure is the end of hypertrophic cardiomyopathy, the decreased protein Ksuc might be a compensatory performance. (2) There might be species differences between mice and human. Collectively, the relationship between succinylation and cardiovascular disease is still worthy to be deeply studied.
Protein succinylation in metabolic diseases
LC–MS/MS analysis have suggested a tight connection between succinylome and diabetes/hepatic steatosis.169,170 Ksuc of optineurin (OPTN), the autophagy receptor, and sterol-regulatory element binding protein 1c (SREBP1c), the key transcription factor in de novo lipogenesis, have been studied in diabetic retinopathy (DR) and hepatic steatosis respectively in depth. DR is a neural and microvascular complication of diabetes and remains the leading cause of blindness in working-aged people (Fig. 4d).171 Elevated Ksuc level of OPTN at K108 with an unchanged protein stability was found in a treptozotocin (STZ)-induced type 1 diabetes rat model, which results in autophagic flux blockade and accumulation of oxidatively damaged proteins or organelles in the cytoplasm under high-glucose conditions. It is possible to speculate that the added large acidic succinyl group inhibits the function of OPTN as a receptor by affecting PPI. In the study of hepatic steatosis, Guo et al. found that the subunit of nuclear factor-κb (NF-κB)—p50 stabilizes HDAC1 to downregulate SREBP1c Ksuc, which increases SREBP1c stability and aggravate hepatic steatosis.172 The different influence of Ksuc on protein stability in S100A10 and SREBP1c indicate an uncovered complex regulatory mechanism of Ksuc.
Protein succinylation in AD
AD has a close relationship with abnormal brain glucose metabolism and accumulation of plaques and tangles. Nonhistone Ksuc has been found to be closely connected with pathological characteristics of AD. In a TMT labeled MS2-based quantitative proteomic study, researchers found that succinylation of multiple mitochondrial proteins declined, and succinylation of small number of cytosolic proteins increased.173 The largest increases occurred at critical sites of amyloid precursor protein (APP) (K612) and microtubule-associated tau (K311), which are crucial to AD progression.174,175,176,177 Mechanistically, succinylation of APP disrupted its normal proteolytic processing thereby promoting Aβ accumulation and plaque formation. Meanwhile, succinylation of tau promoted its aggregation to tangles and impaired microtubule assembly. This study suggested that targeting succinylation of tau, a star AD drug target, may have therapeutic potential against AD in the future.
Malonylation and its role in human diseases
Protein Kmal and its deacylase SIRT5 was identified by the group of Zhao in 2011 after the discovery of Ksuc through mass spectrometry and protein sequence-database searching.178 They identified 25 peptides in 17 nonhistone proteins in HeLa cells, most of which are metabolic enzymes, including glyceraldehyde-3-phosphate dehydrogenase (GAPDH), NOP2/Sun RNA methyltransferase 2 (NSUN2), PGK1, ENO1, MDH2 and aldolase, fructose-bisphosphate A (ALDOA). As described above, malonyl-CoA is either derived from acetyl-CoA catalyzed by ACCase or malonate catalyzed by acyl-CoA synthetase family member 3 (ACSF3),179,180,181,182 which has a close relationship with glucose and fatty acid metabolism. Besides, malonyl-CoA has been shown to elevate in the skeletal muscle and the liver of OLETF rats and in muscle biopsies of patients with obesity and type 2 diabetes (T2D).183 The malonylation proteome reveals that Kmal and its regulation by SIRT5 were more prevalent in the cytosol than mitochondria, which is opposite to Ksuc with a major distribution in mitochondria.184 Nonhistone protein Kmal has been demonstrated to participate in metabolic-associated diseases, including metabolic syndrome, cardiovascular disease, tumor, and non-metabolic diseases, including inflammation.185 In detail, Kmal can be involved in the regulation of protein–protein interaction, mRNA binding ability, and enzymatic activity via changing protein charge from +1 to −1.
Protein malonylation in tumor
Protein Kmal has been suggested to promote cancer via increasing angiogenesis and F-actin nuclear polymerization. Bruning et al. has built the link between fatty acid synthesis with tumor angiogenesis via mTOR malonylation.186 They demonstrated that knockdown or inhibition of FASN helps interrupting the transformation of malonyl-CoA to palmitate, increasing malonyl-CoA accumulation and mTOR K1218 malonylation. This will in turn downregulate mTOR kinase activity and subsequent phosphorylation of p70S6K/4EBP1, promoting endothelial cells proliferation (Fig. 4e). Metastasis is the most common reason for HCC treatment failure and F-actin structure is known to link with the invasive and metastatic phenotypes of cancer cells.187,188 Recently, Kmal was found to promote lung metastasis of liver cancer via promoting polymerization of nuclear actin.189 In this study, the authors focused on the major actin-binding protein mouse diaphanous 2 (mDia2), which contains nuclear localization sequence (NLS) that contributes a lot to actin nuclear translocation and polymerization. They found that loss of mitochondrial transcription factor A (TFAM), a pivotal regulator of mitochondrial biogenesis, will block TCA cycle and cause the accumulation of cytoplasmic acetyl-CoA and its derivative malonyl-CoA, promoting mDia2 Kmal. Mechanistically, malonylation of NLS in mDia2 facilitates its interaction with importin α1 and subsequent nuclear translocation together with actin. Consequently, polymerization of nuclear actin allows chromatin compaction to be reordered to access genetic loci for transcription or repair.190 In this study, malonylation acts as a sensor to build a bridge between mitochondrial biogenesis and metastasis. It is possible that the influence on PPI of Kmal is due to the changed electrostatic interactions and protein conformation caused by the protein charge state switch.
Protein malonyaltion in inflammation
Pro-inflammatory macrophages, such as those activated by LPS, are highly glycolytic with a disrupted TCA cycle.191 In a recent study, Galván-Peña et al. found that malonylation of a wide variety of proteins occurs in macrophages in response to LPS.192,193 They focused on GAPDH, a critical enzyme in glycolysis, and creatively connect metabolic reprogramming with innate immune response via malonylation. This is the first time that protein Kmal was found in immune cells. In detail, LPS upregulates the catalytic activity of ACC1 to provide more malony-CoA for GAPDH Kmal at K213, leading to the increment of its enzymatic activity and dissociation from TNFα mRNA to promote TNFα translation (Fig. 4f). This study is likely to expand our understanding of underlying processes in infection and inflammation from the view of metabolism and protein acylation, and potentially indicate new therapeutic strategies to limit inflammation in disease.
Protein malonylation in cardiovascular disease
Malonylation of proteins especially IDH2, a key enzyme in the TCA cycle and mitochondria, has been explored in cardiomyopathy. Peoples et al. have developed the cardiomyopathy model by using mice with cardiomyocyte-specific depletion of mitochondrial phosphate carrier (SLC25A3), which causes defective mitochondrial ATP synthesis.194 They discovered a striking pattern of acylome remodeling with significantly increased acetylation and malonylation of mitochondria proteins. IDH2 is observed with both upregulated Kace and Kmal, as well as enhanced enzymatic activity. Kace of IDH2 enhances its enzyme activity through neutralizing the positive charge of target lysine, while Kmal plays an opposite role via imparting a negative charge. Deep study illuminated that the increased IDH2 activity is the competing results of its Kace versus Kmal. The biological function of the observed hypermalonylation in cardiomyopathy is still unclear. How does the different protein acylations coexist and compete with each other is worthy to be studied. In another study of cardiac hypertrophy, lower protein malonylation was observed in both transverse aortic constriction (TAC) induced animal model and angiotensin II induced cell model. Among the identified 330 proteins with Kmal, IDH2 was also found to show a significant decrease of Kmal with enhanced enzymatic activity in cardiac hypertrophy (Fig. 4g).195 In the two studies above, it is consistent that upregulation of IDH2 activity in cardiomyopathy connects with disease progression. Whereas IDH2 malonylation is completely opposite, reflecting delicate changes of malonylation in response to genetic, mechanical or pharmacological stresses.
Protein malonylation in metabolic disease
SIRT5 is a NAD+-dependent lysine deacylase responsible for removing malonyl groups. Using affinity enrichment and label free quantitative proteomics, researchers had characterized the SIRT5-regulated lysine malonylome in wild-type (WT) and Sirt5−/− mice. They concluded that SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target, linking Kmal with glucose metabolism.58 Based on their research, another group constructed the hepatic Sirt5-overexpressing ob/ob mouse model to study the biological role of SIRT5 and malonylation in pathological state of obesity.196 They demonstrated that overexpression of Sirt5 results in decreased malonylation and succinylation in hepatic cells, improved cellular glycolysis, suppressed gluconeogenesis, enhanced fatty acid oxidation, and attenuated hepatic steatosis. It is possible that high levels of sirtuins improve the activity of metabolic pathways and reverse some abnormal phenotypes probably by lowering the modification of key metabolic enzymes (Fig. 4h). This study provides an alternative view to understand the mechanism of metabolic abnormality in obesity and T2D, which would benefit for finding novel treatment strategy against these diseases.
Protein crotonylation and its role in human diseases
Kcro has a conjugated double bond that differs from other protein acylation modifications.197 It was first identified in 2011 as an evolutionarily conserved modification in histone proteins via integrated, mass spectrometry-based proteomics approach.197 Six years later, two studies on the identification of nonhistone protein crotonylation have been developed by using the pan lysine crotonylation (α-Kcro) antibody and LC–MS/MS. In one study, 2696 lysine crotonylation sites were identified on 1024 proteins in the human lung adenocarcinoma cell line H1299, in which 40% of crotonylated proteins were in the cytoplasm, 27% were in the nucleus and 13% were in the mitochondria.198 In another study, 453 crotonylated proteins were identified from HeLa cells, in which 62.3% of the proteins were distributed in the nucleus, 9.4% were both distributed in nuclear and cytoplasmic localization, and 28.3% in other cellular components.199 These studies have expanded our understanding of this modification in regulating nonhistone proteins. Besides, it’s possible to speculate that different cell types and disease conditions might accurately decide the different Kcro subcellular distributions. Recently, novel bioinformatics or computational tools for accurate and fast identification of Kcro sites on human nonhistone proteins have been developed, which may serve as an efficient means to assist academicians with their experimental researches.200,201 Histone and nonhistone Kcro have been demonstrated to participate in tumor and ischemic heart disease (IHD) via regulating metabolic enzymatic activity or protein stability.202,203
Protein crotonylation in tumor
In recent years, lysine crotonylation was demonstrated to play opposite roles in diverse cancer types by regulating metabolic enzymes or oncogenic proteins. It is downregulated in liver, stomach, and kidney carcinomas and upregulated in thyroid, esophageal, colon, pancreatic, and lung carcinomas.204 In liver cancer, upregulating the crotonylation level by knocking down HDACs or adding HDAC inhibitors could inhibit hepatoma cell motility and proliferation. However, it helps to promote tumor progression in CRC and cervical carcinoma.
Protein crotonylation regulates glycolysis to aggravate tumor progression
ENO1 is an enzyme in glycolysis that catalyzes glyceric acid-2-phosphate to phosphoenolpyruvate. In addition to its glycolytic function, there is growing evidence to show that ENO1 is an oncogene in CRC tissues.205,206 It was reported that CBP mediates the crotonylation of ENO1 at K420 to promote tumor growth, migration, and invasion of CRC cells in vitro by enhancing ENO1 enzymatic activity and regulating the expression of tumor-associated genes.207
Protein crotonylation in the regulation of oncogenic signaling
Heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1) is highly expressed in a variety of cancers and closely associated with cancer initiation and progression.208,209,210 P300-mediated lysine crotonylation of HNRNPA1 helps to upregulate its protein expression, which in turn promotes HeLa cell proliferation, invasion, and migration.211 However, whether crotonylation hampered HNRNPA1 ubiquitination and degradation to enhance its protein stability still needs to be investigated.
Protein crotonylation in cardiovascular disease
Myocardial ischemia-reperfusion injury contributes to adverse cardiovascular outcomes after myocardial ischemia or cardiac surgery. A proteomic analysis revealed that cardiac ischemia-reperfusion injury causes Kcro of proteins associated with cardiomyocyte contractility, resulting in the disruption of cardiomyocyte mitochondrial, sarcomere architecture, and gap junction, as well as the induction of cardiomyocyte apoptosis.203 The pathological role of Kcro modification on cytoskeletal protein tropomyosin alpha-1 chain (TPM1) and metabolic enzyme IDH3a were further investigated. In detail, Kcro mimicking mutants of IDH3a (K199Q) and TPM1 (K28/29Q) not only protect cardiomyocyte from apoptosis by inhibiting Bcl-2 adenovirus E18 19-kDa-interacting protein 3 (BNIP3)-mediated mitophagy or cytoskeleton structure rearrangement but also preserves post-injury myocardial function by inhibiting fibrosis and apoptosis. However, how does the hydrophobic crotonyl group regulate the biological function of IDH3a and TPM1 is still worth to be studied.
β-hydroxybutyrylation and its role in human diseases
Ketone bodies, including β-OHB, acetoacetate, and acetone, are intermediate metabolites during fatty acid oxidation.212 In the year of 2016, Xie et al. identified a new type of histone modification—Kbhb. They demonstrated that Kbhb is significantly induced during prolonged fasting in mouse liver and is associated with genes upregulated in starvation-responsive metabolic pathways.9 Koronowski et al. also found that dietary and disease states of ketogenesis evoke Kbhb across the cellular proteome.42 Kbhb occurring on histone proteins plays important roles in liver cancer and major depressive disorder.213,214 Studies on nonhistone proteins’ Kbhb are limited and still ongoing.
Protein β-hydroxybutyrylation in tumor
In a study of tumor and diet, researchers found that β-OHB produced by ketogenic diet with low carbohydrate and high fat inhibits CRC development.215 On the contrary, β-OHB treatment induced Kbhb of p53 at K120, K319, and K370 by CBP/p300 to attenuate the cell growth arrest and apoptosis-inducing functions of p53.216 Mechanistically, p53 kbhb results in reduced p53 acetylation and inhibits the expression of p21 and PUMA, two downstream target genes of p53. In fact, ketone bodies, such as β-OHB, are reported to play double-sword roles in cancer biology.217,218 Attenuation of p53 activity upon β-OHB-induced Kbhb might partially explain the complex role of ketone bodies in cancer. The inconsistency of these studies may be because that p53 mutation has not been taken into consideration. The above evidence also reminds us that specific diet intervention or metabolite supplementation should be carefully considered, especially for cancer patients.
Lactylation and its role in human diseases
Studies over the last few years have documented that lactate is not only the end product of glycolysis, but also the major circulating energy source and the regulator of multifunctional signaling molecules in human diseases, especially tumors and inflammation.219,220 For example, lactate has been reported to promote tumor progression by helping cancer cells resist the stress of glucose deprivation, reducing immune surveillance of CD8+ T cells or NK cells, and creating an acidic tumor microenvironment that benefits tumor metastasis.221,222,223,224 Besides, Synovial lactate is a promising biomarker to distinguish septic from aseptic arthritis.225 However, the underlying mechanism is still unclear. In 2019, lactate-derived protein Klac was identified as a novel acylation happened on histone proteins by Zhang et al.12 They demonstrated that the abundance of Klac is mainly determined by the concentration of lactate and the activity of glycolysis, providing a novel possible working mode of lactate. Klac of histone proteins has been associated with lung myofibroblasts, AD and tumor including CCRC, liver cancer, and non-small-cell lung cancer (NSCLC).226,227,228,229,230,231
With the progress of analytical chemistry, numerous nonhistone Klac have been identified, expanding beyond its transcriptional regulation function. Utilizing the signature cyclic immonium (CycIm) ion of Klac generated from the linear immonium (LinIm) ion during MS/MS, Wan et al. has developed a straightforward approach to enable the confident identification of Klac by LC–MS/MS. They unveiled a widespread lactylation proteome beyond histones in both cytoplasm and nucleus from not only the enriched lactylproteome but also the existing unenriched human proteome resources.232 With the help of other chemical tools such as alkynyl-functionalized bioorthogonal chemical reporter (YnLac), nonhistone Klac has also been reported by other studies in recent years.233,234 The limited studies on nonhistone Klac have indicated its critical role in tumor, polymicrobial sepsis and retinopathy.
Protein lactylation in tumor
Based on the Warburg effect, the lactate concentration in tumor tissue is 5–20 times higher than in normal tissue (1.8–2.0 mM), which forms the microenvironment with low pH to promote tumor progression. In the previous study, histone Klac has been suggested to promote tumor development through promoting the immunosuppressive role of tumor-infiltrating myeloid cells or driving TAMs polarization to an M2-like phenotype. Recently, Gu et al., discovered that lactate enhances Treg cells through lactylation of MOESIN at the K72 residue in the cytoplasm, which upregulates TGF-β signaling via TGF-βRI and increases expression of FOXP3. MOESIN is primarily expressed in the cytoplasm, while TGF-βRI is mainly distributed on the cell membrane. The lactylation of MOESIN enhanced the interaction between MOESIN and TGF-βRI via providing an additional hydrogen bond. This study firstly indicated that Klac in nonhistone proteins plays a critical role in PPI and signal transduction. Besides, they found that patients responding to PD-1 treatment have lower levels of MOESIN lactylation than nonresponders, providing a novel idea of targeting Treg cells via anti-lactate approaches in enhancing tumor immunotherapy (Fig. 5).235
Lactate and protein lactylation contribute to the suppressive tumor immune microenvironment. Anaerobic glycolysis produces lactate in the cell cytoplasm, which is transported to the extracellular matrix (ECM) via MCT1/4, resulting in low pH tumor microenvironment (TME). The lactate in the ECM inhibits NK cells tumor infiltration and activity, reduces CD8+ T cells glycolysis, proliferation and cytotoxicity. Lactate also promotes the M2 polarization of macrophages and increases the release of anti-inflammatory cytokines from tumor-associated dendritic cells (TADCs). Besides, lactate enhances Treg cells function through promoting lactylation of MOESIN at the K72 residue in the cytoplasm, which upregulates TGF-β signaling via enhancing the interaction between MOESIN and TGF-βRI and increases expression of FOXP3
Protein lactylation in polymicrobial sepsis
As described above, lactate in the blood has been suggested as a biomarker of sepsis, which is a life-threatening disease characterized by organ dysfunction and dysregulation of inflammatory response.236 High mobility group box-1 (HMGB1) is associated with the severity and mortality of this disease.237 During polymicrobial sepsis, macrophages uptake extracellular lactate through monocarboxylic acid transporter (MCT), promoting HMGB1 Klac in the nucleus through a p300/CBP-dependent manner.237 The lactated HMGB1 will then enhance its acetylation by inhibiting the deacetylase SIRT1 and recruiting the HAT p300/CBP to the nucleus. The lactylated/acetylated HMGB1 in macrophages is then diffused to and accumulated in the cytoplasm and released through the exosome secretion pathway. This will in turn destroy endothelial integrity and increase vascular permeability, leading to endothelial cell barrier dysfunction and promoting sepsis development. Reducing lactate production in vivo can reduce HMGB1 levels and improve survival outcomes in patients with polymicrobial sepsis. In this study, the working mechanisms of Klac and the relationship between Klac and Kace are still indistinct. The molecular weight of HMGB1 is about 25 KD, which shuttles between cytoplasm and nucleus through passive transport. We can hazard guess that the protein polarity change caused by lactyl group might prevent HMGB1 from entering the nucleus.
Protein myristoylation and its role in human diseases
Four kinds of protein lipidation, myristoylation (C14), palmitoylation (C16), prenylation (C15 or C20), and the glycosylphosphatidylinositol anchor, have been identified to be essential in regulating the structure and function of numerous proteins. Among them, myristoylation and palmitoylation are very important in the progress of many diseases.
Since the discovery of 3’,5’-cyclic adenosine monophosphate (cAMP) myristoylation in 1982, approximately 100 kinds of myristoylated proteins have been identified, including serine/threonine kinases, tyrosine kinases, substrates of kinases, phosphatases, regulators of vesicle transport, and signal transduction proteins.238 Although the ratio of myristic acid to total fatty acids is very small, proteins still utilize this kind of rare fatty acid as an N-terminal glycine or lysine modifier of new peptides due to its moderate hydrophobicity. The hydrophobic myristoyl group ensures the proper functioning and intracellular trafficking of proteins. For a long time, the myristol moiety is considered insufficient for protein-membrane associations unless additional membrane-affinity motifs, such as a stretch of basic residues and a second lipid modification. Until recently, Xiong et al. demonstrated that the electrically neutral N-terminal fragment of the protein kinase A catalytic subunit (PKA-c), in which myristoylation is the only functional motif sufficient for membrane association, providing a revised concept for the myristoylation working mode.214 Protein myristoylation has been reported in tumor, immune response, viral infection, and neurological disorders.239
Protein myristoylation in tumor
Protein N-myristoylation has been reported to promote tumor progression through regulating autophagic flux, protein affinity with cell membrane and protein degradation.
Myristoylation regulates autophagy by affecting protein affinity with lysosomes or mitochondrial membranes
AMP-activated catalytic subunit alpha-1 (AMPK) plays critical roles in the surveillance of mitochondrial damage and mitophagy induction.240,241,242 NMT1 mediates AMPKβ myristoylation to enhance its recruitment to damaged mitochondria. This in turn promotes the interaction of AMPK and the ATG16 complex and mitochondrial removal through autophagy, which helps to maintain cancer cell viability in ccRCC, lung cancer, and ovarian serous cystadenocarcinoma (OSC).243 In another two studies, researchers found that myristoylation of the lysosomal adaptor mTOR activator 1 (LAMTOR1) by NMT1 at G2 promotes its palmitoylation and anchoring to the lysosome, which increases mTOR activity to inhibit autophagy and promote the progression of lung cancer, bladder cancer, colorectal cancer, and cervical carcinoma.244,245
Myristoylation affects oncoprotein affinity with cell membranes and their signal transduction capacity
Src is a nonreceptor tyrosine kinase that acts as key mediators of cellular signal transduction and membrane binding. NMT1 regulates Src kinase myristoylation and phosphorylation, which allows the attachment of Src to the cytoplasmic membrane and mediates its kinase activity and cellular functions. Activation of SRC increases many downstream signaling pathways to facilitate tumor growth, angiogenesis, and metastasis in CRC and PCa.246 FGF/fibroblast growth factor receptor (FGFR) signaling is also regulated by myristoylation. For the activation of FGF/FGFR signaling, a scaffold protein called fibroblast growth factor receptor substrate 2α (FRS2α) must be recruited to initiate downstream signaling.247 Myristoylation of FRS2α is essential for the anchoring of FRS2α to the plasmatic membrane, which facilitates its binding with FGFR and helps to activate downstream phosphatidylinositol 3-kinase/AKT signaling and RAS/ERK signaling.248 Activated FGF/FGFR signaling ultimately promotes cell proliferation and migration in several cancer cell types.249,250 For AKT, myristoylation not only enhances its anchoring to the plasma membrane but also confers it to higher basal kinase activity, which upregulates aerobic glycolysis and fatty acid metabolism to promote tumor progression.251,252 Highly metastatic ovarian cancer cells have higher expression of the genes involved in fatty acid metabolism, especially acyl-CoA synthetase long-chain family member 1 (ACSL1), which contributes to an increase in phospholipids with relatively short FA chains, including myristic acid. Abundant myristic acid induces the myristoylation of protein kinase AMPK and Src, which in turn promotes ovarian cancer metastasis.253 These studies indicate that there may exist a positive feedback loop between lipid metabolism and myristoylation to accelerate tumor progression.
N-myristoylation affects oncoprotein degradation
N-myristoylation plays critical roles in boosting liver tumorigenesis, possibly by interfering with the balance between two categories of proteins, N-myristoylation downregulated proteins (NDP, including LXN, RPL29, and FAU) and N-myristoylation upregulated proteins (NUP, including AHSG, ALB, and TF), which oppositely control transformative phenotypes. The two types of proteins were revealed to be negatively and positively regulated by NMT1, respectively. Mechanistically, N-myristoylation influences the ubiquitination of NUP and NDP by affecting their binding with the E3 ubiquitin ligase HIST1H4H.254 Breaking N-myristoylation might be helpful to treat liver cancer.
Protein myristoylation in infectious disease
Recent studies have revealed that protein N-myristoylation is involved in host defense against viral and microbial infections.255 Here, we mainly discussed the former one. Protein N-myristoylation is necessary for viral invasion, assembly, intracellular host interactions and budding apart from a few exceptions.256 For example, the myristoylation of virus-encoded protein (VP0) in rhinoviruses and the gag or nef protein in HIV-1 is vital for viral replication, assembly, and infection257,258,259 The high cell membrane affinity or changed protein activity caused by the myristoyl moiety contributes a lot to these biological processes. As virus is lack of NMT1/2 to catalyze this modification, their proteins are myristoylated by host NMTs. NMT inhibitors such as IMP-1088 have shown pretty good effects on inhibiting virus assembly and replication.258 It is interesting and reasonable to study whether there is myristoylation in proteins of SARS-CoV-2, which may constitute a target in the development of COVID-19 therapeutic drugs.
Protein myristoylation in neurological disease
Parkinson’s disease (PD) is a chronic, progressive neurodegenerative disorder with no satisfied treatment strategy. Virus-delivered neurotrophic protein molecules within brain extracellular space to directly activate the intracellular signaling pathways is a promising therapeutic direction. Akt signaling maintains cell viability through antiapoptotic effects and mediates effects on axonal caliber, branching, and regeneration in neuron.260,261 As described above, myristoylated AKT could be recognized as its constitutive active form. In an in vivo study, Ries et al. demonstrated that transduction of myristoylated AKT via adeno-associated virus confers almost complete protection against apoptotic cell death in a highly destructive neurotoxin model.262
Protein palmitoylation and its role in human diseases
Protein palmitoylation is a reversible dynamic process that was initially discovered in the 1980s. Similar to myristoylation, palmitoylation mainly acts as a lipid anchor that links proteins with specific membrane domains or lipid rafts, which helps signal transduction. In addition, S-palmitoylation is also reported to affect PPIs, protein stability, and protein aggregation. Over 300 proteins have been biochemically confirmed to be S-palmitoylated, and many of them function in cancer, COVID-19, neurological disease, immune disease, and cardiovascular disease.263,264,265,266,267,268,269,270
Protein palmitoylation in tumor
Palmitoylation regulates tumor glycolysis
Glucose transporter (GLUT1) is a transmembrane protein that is responsible for the uptake of glucose into cells through facilitative diffusion and has been demonstrated to promote tumor progression in many cancer types.271,272,273,274 Plasma membrane (PM) localization is essential for glucose uptake by GLUT1. Researchers found that ZDHHC9 mediates the palmitoylation of GLUT1 at C207 and promotes its PM localization process, leading to a high level of glycolysis and glioblastoma (GBM) tumorigenesis.275
Palmitoylation regulates ER stress, and ER-controlled calcium (Ca2+) signal homeostasis
Disturbances in endoplasmic reticulum (ER) homeostasis activate the ER stress response, which promotes tumor cell apoptosis. Protein kinase R-like ER kinase (PERK), inositol-requiring enzyme 1a (IRE1a), and activating transcription factor-6 (ATF6) are three proteins that mediate the ER stress response.276,277,278 X-box binding protein 1 (XBP1) is a key transcription factor in the IRE1a signaling branch and is spliced by endoribonuclease and IRE1a to regulate protein folding, trafficking, and secretion, thus enhancing cell survival and ER homeostasis under ER stress.279 Researchers found that most PATs are upregulated in GBM. Inhibition of palmitoylation using 2-bromopalmitate (2-BP), cerulenin, and tunicamycin induces ER stress and cell death by inhibiting XBP1 palmitoylation. Ca2+ is a secondary messenger in cells and is mainly stored intracellularly in the ER to sustain a nanomolar cytosolic level. Malignant tumor cells usually exhibit a strong dependence on cytosolic Ca2+ for disease progression.280,281,282 Inositol 1,4,5-trisphosphate (IP3) interacts with IP3R in the ER membrane to release Ca2+ from the ER lumen.283,284 Fredericks et al. found that selenoprotein K (SELENOK) forms a complex with ZDHHC6 on the ER membrane and promotes IP3R palmitoylation at C56 and C849, leading to IP3R protein stabilization and Ca2+ release. By using spontaneous metastatic melanoma model with conditional SELENOK knockout mice, Michale et al. demonstrated that deleting SELENOK reduced tumor stemness, tumor growth and metastasis. This may be due to the inhibition of IP3R palmitoylation and reduced calcium flux necessary for tumor growth and metastasis.285
Protein palmitoylation in the regulation of oncogenic signaling
The protein palmitoylation of key oncoproteins helps to promote or inhibit tumor progression. The membrane protein claudin-3 (CLDN3) is critical for the formation and maintenance of tight junctions and is highly expressed in numerous cancers.286 ZDHHC12 mediates CLDN3 S-palmitoylation on three juxtamembrane cysteine residues (C181, C182, and C184), which contribute to its accurate localization in plasma membranes and protein stabilization, promoting ovarian cancer tumorigenesis. Palmitoylation also regulates G-protein-coupled protein activity.287 RAS proteins transduce extracellular signals from activated receptors at the PM to the nucleus, promoting cell proliferation, metastasis, and survival in ~30% of all human cancers. RAS family members, including NRAS, HRAS, KRAS4A, and KRAS4B, are highly homologous but distinct in the -CAAX motif at the C-terminus, which is subjected to PTMs to deliver RAS from the Golgi to the PM.288 Farnesylated and palmitoylated RAS proteins have a 100-fold higher affinity with membrane than that of only farnesylated Ras proteins. Researchers have demonstrated that the palmitoylation of oncogenic NRAS mediated by ZDHHC9 is essential for its PM localization and downstream signaling activation, promoting leukemogenesis.289,290 Epidermal growth factor receptor (EGFR) is an essential driver of oncogenic signaling. ZDHHC20 palmitoylates EGFR on the C-terminal domain (C1025) to promote its ubiquitination-mediated lysosomal degradation, thus upregulating p85 binding and phosphatidylinositol 3-kinase (PI3K) signaling and inhibiting Grb2 binding and MAPK signaling. By using a KrasG12D-induced spontaneous lung cancer mouse model and Zdhhc20 knockout mice, researchers demonstrated that the ablation of ZDHHC20 or induction of EGFRC1025A mutant inhibited tumorigenesis, which seems contradictory to the increased EGFR activation. They explained that EGFR palmitoylation appears to be necessary for PI3K-AKT-Myc signaling in lung cancer, revealing that ZDHHC20 inhibition represents a therapeutic vulnerability in the PI3K-AKT pathway.291 Melanocortin-1 receptor (MC1R) is a key protein in determining hair and skin pigments. MC1R mutations often results in the occurrence of red hair color and promotes melanoma tumorigenesis under ultraviolet (UV) irradiation.292,293 Researchers have demonstrated that palmitoylation of MC1R mediated by ZDHHC13 under UV irradiation helps to enhance DNA damage repair and inhibit melanoma genesis. This is the only study in which the palmitoylation of proteins was demonstrated to inhibit tumor progression.294
Protein palmitoylation in shaping antitumor immune responses
Many palmitoylated proteins have a close relationship with the immune system. In a recent review,295 the authors summarized the important immune signaling pathways associated with palmitoylation, including STING, NOD1/2, and JAK-STAT in cytokine signaling, T-cell receptor signaling, chemotactic GPCR signaling, apoptosis, phagocytosis, and endothelial and epithelial integrity. Studies about protein palmitoylation in the regulation of tumor immune microenvironment is also emerging, mainly focusing on PD-1/PD-L1 and IFN-γ signaling. PD-L1 was found to be palmitoylated at C272 by ZDHHC9 in breast cancer to enhance its stability. Disrupting PD-L1 palmitoylation by site-specific point mutation or inhibiting the expression of its PAT ZDHHC9 sensitized breast cancer cells to T-cell killing and thus repressed tumor growth.296 Another group formulated the same conclusion with more details in CRC. They found that the palmitoylation of PD-L1 increases its binding with PM and inhibits its ubiquitination-mediated lysosomal degradation, while the PAT is ZDHHC3 but not ZDHHC9 in CRC.297 The same group further illuminated the regulatory role of palmitoylation on PD-1 stability in tumor cells. Mechanistically, ZDHHC9 mediates PD-1 palmitoylation at C192 to promote its binding with recycling endosomes, thus preventing its lysosome-dependent degradation. They demonstrated that palmitoylation of PD-1, but not PD-L1, promoted mTOR signaling and tumor cell proliferation.298 IFN-γ belongs to the type II interferon family and is secreted by activated immune cells. IFN-γ signaling plays a critical role in antitumor responses, as it activates the Janus kinase (JAK) signal transducer and activator of transcription 1 (STAT1) pathway to induce the expression of classical interferon-stimulated genes that have key immune effector functions.299 recent work found that C122 palmitoylation in IFN-γ receptor 1 (IFNGR1) acts as a sorting signal for IFNGR1 lysosomal degradation mediated by AP3D1 in CRC. OPTN, which is lost in early-stage human colorectal cancer, interacts with AP3D1 to hinder its recognition of IFNGR1, thereby maintaining IFNGR1 stability and the integrity of downstream MHC-I signaling, which promotes antigen presentation to T cells and inhibits CRC progression (Fig. 3c).300 This work suggests that targeting IFNGR1 palmitoylation might be a new strategy for promoting immune checkpoint therapy in CRC.
Protein palmitoylation in COVID-19
During the SARS-CoV-2 infection, it’s a key event that the spike (S) protein in the virus directly interacts with the receptor angiotensin-converting enzyme 2 (ACE2) in the host cell surfaces. Recently, two studies have separately reported the palmitoylation of both S protein and ACE2 protein. Both the C15 and cytoplasmic tail of S protein were palmitoylated by ZDHHC2, 3, 4, 5, 8, 9, 11, 14, 16, 19, and 20, which is critical for S‐mediated syncytia formation and SARS‐CoV‐2 pseudovirus particle entry.301 In another study, the authors discovered the presence of ACE2 in extracellular vehicles (EVs) which are determined by protein palmitoylation. The C141 and C498 residues on ACE2 are palmitoylated by ZDHHC3, which is critical for the membrane-targeting of ACE2 and their EV secretion.302 Besides, they built the engineered PM-ACE2-EVs via fusing the S-palmitoylation-dependent PM targeting sequence with ACE2 to bind with SARS‐CoV‐2, which blocks S protein interaction with ACE2 and protect host against SARS-CoV-2-induced lung inflammation. It’s reasonable to speculate that ZDHHC inhibitors might also be a potential therapeutic strategy in treating COVID-19, which might kill two birds with one stone.
Protein palmitoylation in neurological diseases
Protein palmitoylation is an important process to regulate the physiological function of the brain, which plays important roles in AD, Huntington disease (HD) and depression. We have mentioned above that protein Ksuc of APP is critical to Aβ production. Here, researchers found that the palmitoylation of APP cleaving enzyme (BACE1) also positively determines the formation of Aβ, resulting in cognitive decline. This modification mainly decides dendritic spine localization and axonal targeting of BACE1 but not its enzymatic activity or protein stability.303 Besides BACE1, palmitoylation of γ-secretase complex, Fyn and flotillins/reggies have also been linked with AD progression.303 HD damages the corticostriatal circuitry in large part by impairing transport of brain-derived neurotrophic factor (BDNF), which is dependent on the protein of huntingtin.304 Pamitoylation of huntingtin by HIP14 (ZDHHC17) is essential for its trafficking and function.305,306 Restoring brain palmitoylation by inhibiting APT1 with the brain-permeable and selective molecule ML348 rescues neuron trafficking, which alleviate behavior and neuropathology of HD mice.307 Besides neurodegenerative disease, key proteins in depressive disorder is also found to be regulated by palmitoylation, including serotonin 1A receptor (5-HT1AR) and postsynaptic density protein 95 (SD-95).308,309 These studies provide promising clinical strategies for the treatment of neurodegenerative diseases and depression via manipulating palmitoylation of specific proteins. Protein palmitoylation and myristoylation both regulate tumor, infectious diseases and neurological diseases through affecting protein-membrane binding, therefore we summarized them in Fig. 6.
Protein palmitoylation and myristoylation within tumor, infectious diseases and neurological diseases in affecting protein-membrane binding. In the protein palmitoylation process, DHHC domain of ZDHHCs binds to the palmitoyl-CoA located in the membrane and undergoes autopalmitoylation, which is followed by a transfer of the palmitate group to the cysteine residue of the substrate protein, promoting the membrane localization of the substrates. The myristoylation process is similar with palmitoylation. First, NMT binds the fatty acid chain of myristoyl-CoA to form the myristoyl-CoA-NMT complex accompanied by substrate-binding pocket exposure, allowing the substrate protein to bind. Second, the NMT catalyzes N-myristoylpeptide formation through chemical transformation and releases the myristoylpeptide and CoA. These two kinds of PTMs promote tumor progression through regulating oncogenic signaling pathways, autophagy, tumor metabolism, ER stress, and tumor immune microenvironment. Besides, they are also critical to the infection of COVID-19, Rhinovirus and HIV, and play important roles in neurological diseases, including Alzheimer’s disease (AD), Huntington’s disease (HD), Parkinson’s disease (PD), and depression
Crosstalk between metabolism and acylation in human diseases
Generally speaking, nearly all kinds of acylation respond to changes in metabolism, which suggests that acylation may help to integrate the responses to metabolic challenges. Both cell-intrinsic tumor metabolic reprogramming and cell-extrinsic factors, including dietary structure, dietary supplement and metabolic microenvironmental factors, such as obesity, diabetes and low pH, can regulate the acylation of proteins. Conversely, protein acylation reactivates and introduces changes to metabolism. Here, we summarized the topic from the following two aspects: how intrinsic and extrinsic metabolic state regulates protein acylation, and how protein acylation reciprocally regulates metabolism (Fig. 7). We mainly focus on tumor as examples.
Crosstalk between protein acylation and metabolism in human diseases. Protein acylation on the metabolic enzymes or transporters always modulate human diseases through affecting glycolysis, fatty acid, and amino acid metabolism. As described in the left, LDHA succinylation, LDHB, ENO2, and PGK1 acetylation, GAPDH malonylation, ENO1 crotonylation and GLUT1 palmitoylation (Spal) can regulate glycolysis. SHMT2 succinylation and IDH1 acetylation regulate amino acid (AA) metabolism. Besides, GNPAT acetylation, SREBP1c, ECHA, and PKM2 succinylation influence fatty acid metabolism. From the view of disease, succinylation of LDHA, SHMT2, acetylation of ENO2, PGK1, IDH1, and GNPAT, crotonylation of ENO1 and palmitoylation of GLUT1 are critical in tumor; acetylation of LDHB and succinylation of SREBP1c is critical in hepatic steatosis; malonylation of GAPDH and succinylation of PKM2 is critical in inflammation; succinylation of ECHA is critical in cardiovascular disease. As described in the right, cell intrinsic and extrinsic metabolism conditions will in turn affect the eight kinds of protein acylations
The intrinsic and extrinsic metabolic state on protein acylation
From the view of cell-intrinsic metabolic state, glycolysis, lipid β-oxidation, amino acid catabolism, and metabolism in TCA cycle all contribute to protein acylation. High cellular glucose uptake produces plenty of acetyl-CoA molecules and usually promotes protein acetylation especially in tumor.101,310,311 Lactate, a glycolytic product and a significant energy source also contributes a lot to protein acylation. Lactate could shuttle both intracellularly and intercellularly, establishing metabolic communication between different cell types in the microenvironment. For example, excessive lactate produced by both tumor and stromal cells can shuttle to Treg cells to induce lactylation of MOESIN at K72, which enhances tumor progression via sustaining Treg function.235 Lipid β-oxidation in mitochondria or the cytoplasm can enhance the abundance of acetyl-CoA, crotonyl-CoA, β-hydroxybutyryl-CoA, myristoyl-CoA and palmitoyl-CoA to promote the corresponding modifications. In addition, amino acid catabolism contributes greatly to acetyl-CoA and crotonyl-CoA for acetylation and crotonylation. High levels of α-KGDHC in the TCA cycle provide ample succinyl-CoA molecules for succinylation. Besides the supply of acyl groups, non-acyl metabolites have emerged as important regulators of acylation. The dinucleotide NAD+, a key metabolite that is involved in cellular electron transfer reactions, is a required cofactor for sirtuin. NAD+ mediates deacetylation, demalonylation and desuccinylation of target proteins.312,313 The NAD+/NADH ratio reflects the cellular redox homeostasis, suggesting that protein acylation could also act as a sensor to convert redox state to different cellular signaling. It is predictable that more details of metabolites in the regulation of nonhistone protein function will be elucidated to describe the complex interaction between acylation and metabolism.
Regarding extrinsic cell metabolism, diabetes and obesity or high-glucose/high-fat diet, ketogenic diet, starvation, and dietary supplement have been demonstrated to aggravate tumor progression by regulating protein acylation. Glycolysis produces acetyl-CoA, which is the donor of protein acetylation. Our recently published study has demonstrated that CRC patients with diabetes exhibit higher expression of p300 and TRIB3 than that of nondiabetic patients. Mechanistically, the high-glucose tumor microenvironment caused by diabetes mellitus could provide more acetyl-CoA and upregulate TRIB3 acetylation and protein stability, transforming metabolic signals to oncogenic signals.101 It seems reasonable to speculate that diabetes-associated hyperglycemia and high-glucose diet might promote tumor development by upregulating the protein acetylation described above. Lipid metabolism contributes greatly to protein acetylation, crotonylation, myristoylation, palmitoylation and β-hydroxybutyrylation. Obesity-associated high protein expression of leptin and TGF-β has been demonstrated to upregulate SMAD2 acetylation by inactivating ACC1 and promoting breast cancer metastasis.314 The donors of myristoylation and palmitoylation, myristic acid, and palmitic acid mainly exist in natural vegetable oil. Protein myristoylation and palmitoylation have been demonstrated to promote tumor progression by inhibiting autophagy and ER stress, activating numerous oncogenic signaling pathways and regulating immune response-related proteins. Ketogenic diet, fasting or calorie restriction could induce increased levels of β-OHB, which binds to HDAC1, HDAC2 and HDAC3 to inhibit their activities. Based on the HDACs inhibitory role, β-OHB can not only induce Kbhb modification, but also other types of acylation, such as acetylation.315,316,317 Therefore, limiting oil intake and preventing excessive hunger might help to prevent and even inhibit tumor growth. Mitochondrial adaptation is also crucial for the response to calorie restriction.318 In calorie-restricted mice, dramatic changes in acetylation were observed in liver mitochondria, with the majority changes of acetylation increasing. By contrast, in brown adipose tissue, calorie restriction led to markedly decreased mitochondrial acetylation. The strikingly different acetylation change between the two tissues may reflect the different physiological function of liver and brown adipose in response to calorie restriction. Dietary supplement also regulates protein acylation. L-carnitine, sphingosine-1-phosphateand inositol phosphates are also known to inhibit or activate the deacetylase activity of class I HDACs, resulting in histone acetylation and epigenetic regulation of gene expression.319,320,321
Protein acylation in the regulation of metabolism
As described above, PGK1 is acetylated by PCAF at K323 to promote glucose uptake and liver tumorigenesis. ENO2 is deacetylated by HDAC3 at K394 to increase its activation and enhance glycolysis, resulting in PDAC metastasis. LDHA and ENO1, two critical enzymes of glucose anaerobic oxidation, are succinylated and crotonylated by CPT1A and CBP, respectively, to upregulate the production of lactate, aggravating tumor development. In addition, the glucose transporter GLUT1 is palmitoylated by ZDHHC9 to enhance glycolysis, providing energy for tumor progression. GNPAT is acetylated by ACAT1 at K128 to inhibit FASN degradation and enhance lipid synthesis. The IDH1 involved in glutamine metabolism is deacetylated at K224 to inhibit its enzymatic activity and the HIF1α-SRC transcription axis. SIRT5 catalyzes the desuccinylation of the serine metabolic enzyme SHMT2 to regulate one-carbon metabolism and promote tumor proliferation. c-Myc increases SDHA acetylation at K335 by promoting SKP2-mediated SIRT2 degradation to promote succinate production in the TCA cycle and H3K4me3 activation. In summary, protein acylation of metabolic enzymes or nutrient transporters always modulate the progression of different diseases by regulating glucose, fatty acid and amino acid metabolic reprogramming.
Crosstalk among protein PTMs
Crosstalk among protein acylations
Lysines can be posttranslationally modified by various types of acylations. A systematic study showed that a majority of succinylation sites in bacteria, yeast, and mouse liver were acetylated at the same position.156 It is predictable that one lysine site could be modified by several kinds of acylations. These modifications will form a competition with each other. Notably, except for lipidation, all the short-chain protein acylations share common regulatory enzymes, including writers, erasers and readers. For example, p300 almost mediates all the acylation of short-chain protein acylations. SIRT5 is simultaneously responsible for the deacylation of Kace, Ksuc, Kmal, and Kglu. However, the acylations of substrates are not often concurrently change with the regulatory enzymes. As described above, SIRT5 plays critical roles in cardiac function. Depletion of SIRT5 in transgenic mouse enhances protein succinylation significantly but has nearly no influence on protein acetylation.40 It is rational to guess that there are some preconditions for deciding enzymes’ selectivity in catalyzing their substrates, such as the different concentrations of acyl-CoA in various tissues, and the different binding affinity of enzymes with acyl-CoA.
Crosstalk between acylation and other PTMs
Most mammalian proteins are modified by multiple PTMs, such as ubiquitination, phosphorylation, methylation and so on. These PTMs can reciprocally influence each other. Take acetylation as an example, Kace crosstalk with other PTMs can be principally classified into antagonistic or cooperative forms. For the antagonistic crosstalk, sometimes different modifications compete the same lysine sites with acetylation. The E3 ligase E4F1 and the acetyltransferase PCAF mediate mutually exclusive post-translational modifications of ubiquitination and acetylation of p53 at K320, respectively, in the determination of alternative cell fates: growth arrest or apoptosis.322 Also, the antagonistic crosstalk could happen at different amino acid sites. Activation by kinase-mediated phosphorylation and attenuation by phosphatase-mediated dephosphorylation are hallmarks of STAT1 signaling. Acetylation of STAT1 at K410 and K413 facilitate the association of p-STAT1 with phosphatases TCP45, allowing the dephosphorylation of STAT1 and adjust cytokine-induced gene expression rapidly and economically.323 It bears thinking about that under what situation the acetyl group could be added on p-STAT1 to induce it dephosphorylation and deactivation, but not maintaining an active state continuously.
Methylation-acetylation interplay in p53 is an example of cooperative crosstalk. In response to DNA damage, Set7/9 induces K372 methylation of p53, which subsequently enhances p53 acetylation and stabilization.324 Protein N-myristoylation of Src family kinases is critical to anchor the enzymes in the plasma membrane. In fact, it also acts as cooperative signal for Src family kinases phosphorylation. S13 of Lyn (Lyn-S13) could be phosphorylated by casein kinase 1γ (CK1γ) in a Lyn-G2 N-myristoylation-dependent manner at the Golgi during intracellular protein traffic.325 From these examples, we can get the impression that the assortment of different modifications on one protein is nonrandom and may occur in an ordered fashion, allowing precise control of protein stability or activity.
Therapeutic opportunities for targeting protein acylation
Protein PTM has a strong relationship with many human diseases, including metabolic disease, cardiovascular disease, inflammatory and infectious disease, neurological disease, and tumor. Inhibitors or activators targeting protein PTMs have long been developed and showed great prospects in the treatment of those diseases, especially of cancer. For example, five inhibitors of HDACs have already been used to treat hematology in clinic including vorinostat (Merk, USA), romidepsin (Celgene, USA), panobinostat (Novartis, USA), chidamide (Chipscreen, China) and belinostat (Spectrum/Onxeo, USA).326 Many inhibitors of protein acylation against cancer have also been in clinical trials. Therefore, we mainly summarized the potential compounds targeting writers, erasers and readers of protein acylation in the treatment of cancer as follows. Chemicals targeting other diseases in the clinical trials are listed in Tables 2–4.
Development of pharmacological agents for targeting protein acylation writers
Different subtypes of acyltransferases catalyze the acylation of histones and nonhistone proteins on specific amino acid residues. A potential role of the acylation modifications in the pathogenesis of cancer, cardiovascular disease, neurological disease, and immunological or infectious disease has been described. This indicates that specific inhibitors of acyltransferase are potential tools for pharmacological research and might find therapeutic applications.
Therapeutic opportunities for targeting writers of short-chain protein acylation
The acyltransferases of short-chain protein acylation are almost all overlapped and mainly belong to the KAT family except for the specific writers of Ksuc (CPT1A) and Kmal (PMAT1). CPT1A is not only a succinyltransferase but also a protein that catalyzes the rate-limiting step of fatty acid oxidation (FAO). ST1326, the only discovered inhibitor of CPT1A, has been demonstrated to inhibit the proliferation of acute myeloid leukemia. However, it achieves this effect mainly by regulating FAO but not protein succinylation.162,163,164 Besides, there is no compound targeting PMAT1. Therefore, we mainly summarized the inhibitors or activators targeting KAT families. The p300/CBP family is a main component of KATs, which play important roles in regulating the cell cycle, proliferation and differentiation and are highly expressed in colorectal cancer, lung cancer, breast cancer, liver cancer, prostate cancer, and different kinds of leukemia.327,328,329,330,331,332 Researchers have focused their efforts on targeting p300/CBP for a long time, and over 135 compounds have been discovered, which mainly target the HAT domain and BRD domain of these proteins.333,334 Here, we mainly review the compounds of high affinity and selectivity of p300/CBP with satisfactory anticancer effects.
Among the compounds that target the BRD domain of p300/CBP, an orally bioavailable, potent, and selective inhibitor of the p300/CBP bromodomain called CCS1477 was developed by the Cellcentric company. They demonstrated that CCS1477 inhibits cell proliferation in prostate cancer cell lines and decreases AR- and c-Myc–regulated gene expression.331 A phase I/II study to assess the safety, tolerability, pharmacokinetics and biological activity of CCS1477 in patients with metastatic castration-resistant prostate cancer (mCRPC) or advanced solid tumors was started in 2018. Another oral small molecule FT-7051 has been developed by the Forma Therapeutics Holdings, Inc. (FMTX), a clinical-stage biopharmaceutical company. It is designed to attach to the p300/CBP bromodomain potently and selectively, which then blocks androgen binding and reduces AR activation. This compound has shown an encouraging safety and effectiveness in the treatment of metastatic castration-resistant prostate cancer during the phase I clinical trial. EP31670 (NEO2734) is a novel potent oral dual BET and p300/CBP inhibitor, which shows more efficient tumor-killing ability than non-dual BET inhibitors in numerous solid tumors.335 This compound has also entered into the phase I clinical trial in the treatment of castrate-resistant prostate cancer and NUT carcinoma (Table 2).
In compounds that target the HAT domain, C646 and A485 both have high selectivity and antitumor effects. Bowers et al. and shrimp et al. obtained a pyrazolone small molecule compound (C646) through virtual screening, which has been demonstrated to inhibit tumor proliferation, promote tumor apoptosis, alter glucose metabolism, reduce tumor drug resistance and induce tumor immune response in different kinds of cancers.336,337,338,339,340,341,342,343 For example, in our study, C646 was confirmed to inhibit the K240 of TRIB3 and promote CD8+ T-cell recruitment in CRC tissues. The combination of C646 and PD-1/PD-L1 blockade therapy exhibited an increased antitumor effect compared to that of single immune checkpoint therapy.101 Researchers at the Abbvie company designed a class of compounds with the structure of spirindan hydantoin. After further optimization, they finally obtained compound A485 with nanomolar affinity, high selectivity, drug-like properties, and oral bioavailability.344 The compound has been demonstrated to have significant inhibitory activity in NSCLC, mantle cell lymphoma, multiple myeloma, non-Hodgkin’s lymphoma cells, and AR-positive prostate cancer cells.345,346,347 Similar with C646, A485 can enhance the tumor-killing effect of PD-L1 antibody.348 Mechanistically, it blocks the acetylation of histone H3 at the PD-L1 promoter and inhibits its transcription. In a recent study, researchers developed a proteolysis-targeting chimera (PROTAC) compound termed JQAD1 by linking A485 with the CRBN that selectively targets p300 for degradation. JQAD1 has been demonstrated to cause neuroblastoma cell apoptosis concurrent with MYCN downregulation.349 Compounds that target other KAT families are still in the early developmental stage and are not discussed here.
Therapeutic opportunities for targeting writers of protein lipidation
NMT and ZDHHC families are writers for protein myristoylation and palmitoylation. To date, NMT1, NMT2, ZDHHC2, ZDHHC3, ZDHHC6, ZDHHC8, ZDHHC9, ZDHHC12, ZDHHC13, and ZDHHC14 have been closely linked with most cancer types.275,287,350,351,352,353,354,355,356 The development of inhibitors targeting NMT and ZDHHC family to treat cancer is in progress.
The compound used as NMT inhibitors to combat tumors are still at the stage of laboratory research, including 2-hydroxymyristic acid 3, D-NMAPPD (also named B13), Tris-DBA palladium, IMP-366 (DDD85646), IMP-1088 and PCLX-001.258,357,358,359,360 Myristic acid analog 2-hydroxymyristic acid 3 was the first compound reported to inhibit NMT. B13 has been suggested to inhibit tumor progression by downregulating LAMTOR1, SRC and FRS2α myristoylation, as mentioned above.244,246 Tris-DBA palladium has been reported to have antitumor activity against melanoma. However, in a recent study, scientists found that N-myristoylation was unaffected by 2-hydroxymyristic acid 3, B13, and Tris-DBA palladium, and the tumor-killing ability involves some kind of off-target toxicity. IMP-1088, IMP-366, and PCLX-001 (which is orally bioavailable derivatives of IMP-366) have been identified as on-target and selective inhibitors of NMT.359 IMP-366 has been suggested to drive tumor apoptosis and reduce tumor growth by preventing mTORC1 activation and blocking lysosomal degradation. PCLX-001 has been demonstrated to inhibit the growth of hematological cancer cells in vitro and alleviate tumor progression in lymphoma murine xenograft models. Mechanistically, PCLX-001 promotes the degradation of numerous myristoylated and nonmyristoylated BCR effectors, triggering apoptosis. This potential compound has entered the phase I trial in treating B-cell non-Hodgkin lymphoma and advanced solid malignancies, and further tests will be performed361,362 (Table 2).
The development of anti-palmitoylation drug face huge hurdles. Presently, no potent and specific inhibitors against ZDHHC proteins have been discovered.363,364 2-BP, cerulenin, and tunicamycin are PAT inhibitors that are extensively used in the literature, and 2-BP is the most common one. For example, these three compounds have been demonstrated to induce G2 cell cycle arrest and cell death in GBM cells through enhanced ER stress.279 In addition, tunicamycin (5, 10, and 25 μg/ml) and 2-BP (25, 50, and 150 μM) significantly decreased CSC sphere formation in the liver without affecting cell viability.365 2-BP could also activate antitumor immunity in vitro and in mice bearing MC38 tumor cells. However, the nonspecificity of 2-BP limits its development into a drug. Efforts to screen for more selective inhibitors have been made, but this endeavor is difficult due to the extreme similarity of the PATs. Competitive inhibition is an effective approach for targeting specific enzymes. After identifying the palmitoylation motif of PD-L1 and PD-1, Yao et al. synthesized peptides that were derived from PD-L1 (MMDVKKCGIQDTNS-GFP) and PD-1 (Flag-AVICSRAARG-GFP), and these peptides exhibited a successful inhibitory effect that involved PD-1/PD-L1 palmitoylation and slowed tumor progression.297,298 Future studies are necessary to develop more selective PAT inhibitors.
Development of pharmacological agents for targeting protein acylation erasers
Most of the protein acylations are dynamic, reversible, and highly regulated chemical reactions. HDAC family members regulate the homeostasis of protein acylation. Inhibitors of HDACs were used as anticancer drugs or even neuroprotectors by enhancing synaptic plasticity, learning and memory in a wide range of neurological disorders.
Therapeutic opportunities for targeting erasers of short-chain protein acylation
As described above, HDAC family are the common erasers for short-chain protein acylation, participating in tumor progression. Class I HDAC proteins are mainly distributed in the nucleus, among which HDAC1 and HDAC2 are highly expressed in lung cancer, breast cancer, gastric cancer, ovarian cancer, colorectal cancer, and pancreatic cancer.366,367,368,369,370 Class II HDAC proteins are mainly distributed in the cytoplasm and can shuttle from the cytoplasm to the nucleus, and HDAC4 and HDAC6 are highly expressed in liver cancer and oral squamous cell carcinoma, respectively.371,372 Class III contains members of the sirtuin family, which are usually responsible for the deacetylation of nonhistone proteins. SIRT5 and 7 have been demonstrated to promote tumor progression in lung and breast cancers. The only class IV HDAC member, HDAC11, has also been reported to promote lung cancer373 and hepatocellular carcinoma374 in recent years.
Targeting HDAC families is a novel therapeutic strategy against cancer. As described above, four products that target HDACs have been approved by the food and drug administration (FDA) of the USA, including vorinostat, romidepsin, belinostat, and panobinostat for the treatment of cutaneous T-cell lymphoma. Another product has been approved by China food and drug administration (CFDA) for the treatment of peripheral T-cell lymphoma—chidamide (tucidinostat), marking the first example of a “Made in China” drug. Except for chidamide, which is a selective inhibitor of class I HDACs and HDAC10, the other four compounds are all pan HDAC inhibitors. These compounds provide good antitumor effects on leukemia but unsatisfactory effects on solid tumors in the monotherapy. Therefore, clinical trials are currently being performed, in which the combination of HDAC inhibitors with chemotherapeutic drugs, kinase inhibitors or checkpoint inhibitors are being tested for treating solid tumors.375,376 Beyond the five compounds on the market, many novel HDAC inhibitors have entered clinical trials. For example, CRA-024781 (ABEXINOSTAT) with high selectivity of HDAC6 and 8, entinostat with high selectivity of HDAC1 and 3 and the pan HDAC inhibitor CI-994 have all entered the phase III clinical trial for the treatment of renal cell carcinoma, advanced breast cancer, and lung cancer.377,378 Many others are in the phase I–II clinical trials for the treatment of solid tumor or leukemia.379,380,381,382,383 Besides, some compounds showed potential effectiveness in treating neurological diseases, metabolic diseases or inflammatory and infectious disease and are tested in clinical trials (Table 3).
Therapeutic opportunities for targeting erasers of protein lipidation
Although in most cases protein palmitoylation and myristoylation contribute to tumor progression, they also exhibit antitumor activity in some cases. It might be more effective to prevent depalmitoylation or demyristoylation in such specific cases. SIRT6 and IpaJ are erasers of lysine and N-terminal glycine myristoylation, however, no specific inhibitors targeting these erasers to regulate myristoylation have been developed yet. Chemicals targeting APT, PPT, and ABHD families in depalmitoylation are still in their early stage. Palmostatin B (PB) and palmostatin M (PM) are APT inhibitors that covalently modify and inactivate the serine residue in the active site but lack specificity between APT1 and APT2.384,385 PB treatment was found to block the proliferation of AML blasts from a mouse model that harbored oncogenic N-Ras.385 Using fluopol-activity-based protein profiling (ABPP), researchers have identified another APT inhibitor chemotype with a piperazine amide motif and discovered two compounds named Inhibitor 21 and Inhibitor 1, which selectively target APT1 and APT2.386 Beyond inhibition of ATP1/2 to inhibit tumor development, inhibition of PPT1 mediated depalmitoylation may lead to more direct toxic effect in cancer cells. The natural product didemnin B and dimeric quinacrine compounds—DQ661 have been demonstrated to inhibit solid tumor growth via inhibiting PPT1 and disrupting lysosome function.387,388 Except for APT and PPT, inhibitors of the ABHD family are also being developed. A potent and selective covalent inhibitor of ABHD17–ABD957 has been demonstrated to inhibit the proliferation in N-RAS-mutant cancer.389
Development of pharmacological agents for targeting protein acylation readers
BET proteins, especially BRD3 and BRD4, are the readers of nonhistone protein acetylation, which are critical in a wide spectrum of human diseases. More and more evidence supported the use of BETi in treating cancer, metabolic, inflammatory, neurologic, cardiovascular, and musculoskeletal diseases. Over 50 BRD3/BRD4 inhibitors have been developed after the discovery of a first series of compounds in the treatment of cancer, including JQ1, I-BET151, and I-BET762.390,391,392,393,394 These inhibitors can be categorized into two types: (1) inhibitors binding to BD1 or BD2 domains of BRD3/4 and interrupting the enzymatic activity, such as PLX51107 and apabetalone, which have entered the phase I or II clinical trials in the treatment of tumor, COVID-19 infection, cardiovascular disease or diabetes (Table 4).395,396,397 (2) the PROTAC proteins promoting the degradation of BRD proteins, such as BRD4 ligand-Linker Conjugate 1, PROTAC BRD4 degrader-5/15, AT6, OARV-771.398,399 BET inhibitors in monotherapy have shown active effects, while combination studies might have a big impact to improve BET inhibition activity and reduce toxicity.
Conclusions and future perspectives
With the discovery of PTMap, a sequence alignment software for unrestricted, accurate, and full-spectrum identification of PTM sites, research on protein acylation have undergone unprecedented development in the past 10 years. Many novel short-chain protein acylations were identified, which have been deeply studied in various diseases. As all the acylation donors come from metabolites, protein acylation is helping researchers to understand the regulation mechanism of metabolism in both physiological and pathological conditions, providing numerous drug targets in tumor, cardiovascular diseases, metabolic diseases, inflammatory and infectious diseases, etc. This exciting area has become a hot topic in basic research and will bring so many opportunities in translational medicine, however, there are still many challenges to be solved.
Protein acylation in basic research
A lot of efforts have been made in basic research to search for new or specific protein acylations on critical proteins, identify animo acid sites of acylation and study their biological function in different diseases. Several questions or obstacles are existed focusing these work.
-
1.
New approaches besides analytical chemistry of identifying protein acylation are still being needed. Although effective, inaccuracies in spectral matching often result in false-positive identifications in conventional PTM identification via MS, which is based on the fixed mass shift of the modified peptide. The development of complementary approaches is in urgent need. (A) Exploiting signature MS/MS ions to validate the presence of PTMs is invaluable, such as cyclic immonium ion of lactyllysine. (B) Although several pan acylation antibodies were developed and widely used to examine the acylation of proteins in specific cells or tissues, some antibodies targeting specific acylations are still in need, such as the detection antibody for protein palmitoylation. (C) Techniques for the enrichment of acylated peptides may also be useful. For example, scientists reported an enrichment approach based on a novel magnetic microsphere modified with 2,2′-dithiodipyridine (Fe3O4/SiO2-SSPy microsphere), which demonstrated remarkable enrichment selectivity and sensitivity for palmitoylated peptide, enabling a global annotation of protein palmitoylation for complex biological samples.400
-
2.
The methods for verifying protein acylation modification sites need to be carefully considered. Before the discovery of several novel short-chain protein acylations, K to arginine (R) mutation in the substrate was thought to mimic the lysine deacetylation, besides, K to glutamate (E) mutation was thought to mimic the lysine acetylation. However, K-to-E mutation is also considered as a mimic of the succinylated state. Moreover, other PTMs, such as ubiquitination and sumoylation can also occur at lysine residues. That is to say, in some conditions researchers should reconsider whether such mutation really mimics the desired acylation or whether the mutation affects other PTMs of the interest protein, especially in functional study. Although more and more acylation sites were experimentally characterized, the regulatory enzymes for most of sites remain to be dissected. In contrast with labor-intensive and time-consuming experiments, computational prediction of acylation sites from protein sequences based on machine learning is much more helpful to generate highly useful information for further experimental consideration. Several online tools for protein acylation prediction are available now.401,402,403 We have reason to believe that more and more bioinformatic tools will constantly emerge, which will accelerate our comprehensive understanding of protein acylation. Combining of protein acylation prediction and specific PTMs antibody might be helpful for the determination.
-
3.
The donors of several protein acylations have not been found in the human body yet, but from the microbiome in the gastrointestinal tract, including hydroxyisobutyrate, propionate, and butyrate. This might uncovered a new relationship between host and microbiome.
Protein acylation in translational medicine
The discovery of molecular signatures that enables early diagnosis, accurate prognosis and personalized therapy is valuable in clinic, especially in the treatment of cancer. Protein acylation is a dynamic process and has close relationship with tumor, which sheds lights on this valuable requirement. For example, the propionylation level of histone H3K23 in U937 leukemia cells is at least six-fold higher than in non-leukemia cell lines. While, during monocytic differentiation, the propionylation level in U937 cells decreased remarkably, indicating that the initial hyperpropionylation in U937 cells might be a stage-specific marker in leukaemogenesis.404 In another study, authors found that higher levels of H2BK120, H3.3K18, and H4K77 acetylation in liver cancer tissues were significantly associated with worse prognosis, such as poorer survival and higher recurrence in an independent clinical cohort of HCC patients.405 Similarly, nonhistone acylations also demonstrate diagnostic or prognostic potential. High level of acetylated IDH1 at K224 are significantly correlated with advanced tumors, metastasis and reduced survival in CRC.406 Nonhistone acetylation also shows diagnostic potential for infectious disease. Acetylated K676 of transforming growth factor–beta-induced protein (TGFBIp) was consistently elevated in the blood of patients with SARS-CoV-2 pneumonia, especially in patients in the intensive care unit.153 In this study, treatment with TGFBIp neutralizing antibodies suppressed the cytokine storm, suggesting that K676 acetylation of TGFBIp can be used not only as a diagnostic marker, but also a indicator for TGFBIp neutralizing antibody therapy against SARS-CoV-2 pneumonia. For the neurodegenerative disease, succinylated Aβ and tau are closely associated with the disease state of AD,173 providing new molecular diagnostics and potential therapeutic targets. However, several issues are still waiting to be solved in the translational medicine of protein acylations.
-
1.
Large-scale proteomic screening for the acylation of specific proteins in the peripheral blood, fecal or even saliva of patients is crucial to provide more early and convenient diagnostic markers or therapeutic targets for different types of diseases. For this purpose, developing technologies with high specificity and sensitivity to distinguish diverse acylations in a complex system is an urgent issue to be solved.
-
2.
The close relationship of protein acylation and human diseases has rendered it as an attractive therapeutic target. However, there are two limitations in this field. First, the specificity of the inhibitors is generally low. For example, the HDAC inhibitors on the market are almost pan HDACi, producing lots of side effects. Second, the compounds mainly target acyltransferases and deacylases, which have broad influences on downstream substrates. As these enzymes usually play complex roles in regulating different substrates and may act as oncogenic proteins or tumor suppressors in a cancer-type-dependent manner, strategies that target specific protein PTMs are in need. The α-helical interfering peptides might be good candidates to interrupt the association of enzymes with their substrates, providing a more accurate regulation for protein acylation. In addition to the drug therapy, dietary patterns, such as ketogenic diet and restriction of glucose or long-chain fatty acids intake, are also important for the prevention and therapy of human diseases through regulating protein acylation. On this road, the great challenge remains of spatially and timely guiding such interventions to achieve disease-specific outcomes without compromising the responses of healthy cells.
References
Phillips, D. M. The presence of acetyl groups of histones. Biochem. J. 87, 258–263 (1963).
Verdin, E. & Ott, M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 16, 258–264 (2015).
Chen, Y., Chen, W., Cobb, M. H. & Zhao, Y. PTMap-a sequence alignment software for unrestricted, accurate, and full-spectrum identification of post-translational modification sites. Proc. Natl Acad. Sci. USA 106, 761–766 (2009).
Sabari, B. R., Zhang, D., Allis, C. D. & Zhao, Y. M. Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 18, 90–101 (2017).
Tan, M. J. et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 (2011).
Zhang, Z. et al. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 7, 58–63 (2011).
Xie, Z. et al. Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteom. 11, 100–107 (2012).
Tan, M. J. et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 19, 605–617 (2014).
Xie, Z. et al. Metabolic regulation of gene expression by histone lysine beta-hydroxybutyrylation. Mol. Cell 62, 194–206 (2016).
Dai, L. Z. et al. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat. Chem. Biol. 10, 365–370 (2014).
Huang, H. et al. Lysine benzoylation is a histone mark regulated by SIRT2. Nat. Commun. 9, 3374 (2018).
Zhang, D. et al. Metabolic regulation of gene expression by histone lactylation. Nature 574, 575–580 (2019).
Chen, B., Sun, Y., Niu, J., Jarugumilli, G. K. & Wu, X. Protein lipidation in cell signaling and diseases: function, regulation, and therapeutic opportunities. Cell Chem. Biol. 25, 817–831 (2018).
Xu, Y., Shi, Z. & Bao, L. An expanding repertoire of protein acylations. Mol. Cell. Proteom. 21, 100193 (2022).
Hirschey, M. D. & Zhao, Y. Metabolic regulation by lysine malonylation, succinylation, and glutarylation. Mol. Cell. Proteom. 14, 2308–2315 (2015).
Herzig, S. et al. Identification and functional expression of the mitochondrial pyruvate carrier. Science 337, 93–96 (2012).
Rufer, A. C., Thoma, R. & Hennig, M. Structural insight into function and regulation of carnitine palmitoyltransferase. Cell. Mol. Life Sci. 66, 2489–2501 (2009).
Green, C. R. et al. Branched-chain amino acid catabolism fuels adipocyte differentiation and lipogenesis. Nat. Chem. Biol. 12, 15–21 (2016).
Cahill, G. F. Jr Fuel metabolism in starvation. Annu. Rev. Nutr. 26, 1–22 (2006).
Cederbaum, A. I. Alcohol metabolism. Clin. Liver Dis. 16, 667–685 (2012).
Mews, P. et al. Alcohol metabolism contributes to brain histone acetylation. Nature 574, 717–721 (2019).
Metallo, C. M. et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384 (2011).
Schug, Z. T. et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell 27, 57–71 (2015).
Takahashi, H., McCaffery, J. M., Irizarry, R. A. & Boeke, J. D. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol. Cell 23, 207–217 (2006).
Comerford, S. A. et al. Acetate dependence of tumors. Cell 159, 1591–1602 (2014).
Zhang, Y. et al. The pivotal role of protein acetylation in linking glucose and fatty acid metabolism to beta-cell function. Cell Death Dis. 10, 66 (2019).
Matsuzaki, H. et al. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc. Natl Acad. Sci. USA 102, 11278–11283 (2005).
Li, K. et al. Ets1-mediated acetylation of foxo1 is critical for gluconeogenesis regulation during feed-fast cycles. Cell Rep. 26, 2998–3010 (2019).
Liang, D. et al. Alterations of lysine acetylation profile in murine skeletal muscles upon exercise. Front. Aging Neurosci. 14, 859313 (2022).
Lee, J. et al. The acetylation of cyclin-dependent kinase 5 at lysine 33 regulates kinase activity and neurite length in hippocampal neurons. Sci. Rep. 8, 13676 (2018).
Pillai, V. B. et al. Acetylation of a conserved lysine residue in the ATP binding pocket of p38 augments its kinase activity during hypertrophy of cardiomyocytes. Mol. Cell Biol. 31, 2349–2363 (2011).
Sabari, B. R. et al. Intracellular crotonyl-CoA stimulates transcription through. p300-catalyzed histone crotonylation. Mol. Cell 58, 203–215 (2015).
Jeong, J. et al. Energy conservation model based on genomic and experimental analyses of a carbon monoxide-utilizing, butyrate-forming acetogen, eubacterium limosum KIST612. Appl. Environ. Microbiol. 81, 4782–4790 (2015).
Fang, Y. et al. Histone crotonylation promotes mesoendodermal commitment of human embryonic stem cells. Cell Stem Cell 28, 748–763 (2021).
Burch, J. S. et al. Glutamine via alpha-ketoglutarate dehydrogenase provides succinyl-CoA for heme synthesis during erythropoiesis. Blood 132, 987–998 (2018).
Wu, L. et al. Functional characterization of rat glutaryl-CoA dehydrogenase and its comparison with straight-chain acyl-CoA dehydrogenase. Bioorg. Med. Chem. Lett. 21, 6667–6673 (2011).
Sasaki, Y. & Nagano, Y. Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci. Biotechnol. Biochem. 68, 1175–1184 (2004).
Estelmann, S. & Boll, M. Glutaryl-coenzyme A dehydrogenase from Geobacter metallireducens— interaction with electron transferring flavoprotein and kinetic basis of unidirectional catalysis. FEBS J. 281, 5120–5131 (2014).
Park, J. et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell 50, 919–930 (2013).
Sadhukhan, S. et al. Metabolomics-assisted proteomics identifies succinylation and SIRT5 as important regulators of cardiac function. Proc. Natl Acad. Sci. USA 113, 4320–4325 (2016).
Colak, G. et al. Proteomic and biochemical studies of lysine malonylation suggest its malonic aciduria-associated regulatory role in mitochondrial function and fatty acid oxidation. Mol. Cell. Proteom. 14, 3056–3071 (2015).
Koronowski, K. B. et al. Ketogenesis impact on liver metabolism revealed by proteomics of lysine beta-hydroxybutyrylation. Cell Rep. 36, 109487 (2021).
Hou, J. Y., Zhou, L., Li, J. L., Wang, D. P. & Cao, J. M. Emerging roles of non-histone protein crotonylation in biomedicine. Cell Biosci. 11, 101 (2021).
Rampoldi, F. et al. Immunosuppression and aberrant T cell development in the absence of N-myristoylation. J. Immunol. 195, 4228–4243 (2015).
Bijlmakers, M. J., Isobe-Nakamura, M., Ruddock, L. J. & Marsh, M. Intrinsic signals in the unique domain target p56(lck) to the plasma membrane independently of CD4. J. Cell Biol. 137, 1029–1040 (1997).
Rowe, D. C. et al. The myristoylation of TRIF-related adaptor molecule is essential for Toll-like receptor 4 signal transduction. Proc. Natl Acad. Sci. USA 103, 6299–6304 (2006).
Alland, L., Peseckis, S. M., Atherton, R. E., Berthiaume, L. & Resh, M. D. Dual myristylation and palmitylation of Src family member p59fyn affects subcellular localization. J. Biol. Chem. 269, 16701–16705 (1994).
Brigidi, G. S. et al. Palmitoylation of delta-catenin by DHHC5 mediates activity-induced synapse plasticity. Nat. Neurosci. 17, 522–532 (2014).
Shen, Z. C. et al. APT1-mediated depalmitoylation regulates hippocampal synaptic plasticity. J. Neurosci. 42, 2662–2677 (2022).
Kaczmarska, Z. et al. Structure of p300 in complex with acyl-CoA variants. Nat. Chem. Biol. 13, 21–29 (2017).
Kollenstart, L. et al. Gcn5 and Esa1 function as histone crotonyltransferases to regulate crotonylation-dependent transcription. J. Biol. Chem. 294, 20122–20134 (2019).
Liu, X. et al. MOF as an evolutionarily conserved histone crotonyltransferase and transcriptional activation by histone acetyltransferase-deficient and crotonyltransferase-competent CBP/p300. Cell Discov. 3, 17016 (2017).
Wang, Y. et al. KAT2A coupled with the alpha-KGDH complex acts as a histone H3 succinyltransferase. Nature 552, 273–277 (2017).
Gu, L. et al. GNAT-like strategy for polyketide chain initiation. Science 318, 970–974 (2007).
Bao, X. et al. Glutarylation of histone H4 lysine 91 regulates chromatin dynamics. Mol. Cell 76, 660–675 (2019).
Gan, S. et al. The acyltransferase PMAT1 malonylates brassinolide glucoside. J. Biol. Chem. 296, 100424 (2021).
Kurmi, K. et al. Carnitine palmitoyltransferase 1A has a lysine succinyltransferase activity. Cell Rep. 22, 1365–1373 (2018).
Nishida, Y. et al. SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol. Cell 59, 321–332 (2015).
Gong, M., Zhuo, X. & Ma, A. STAT6 upregulation promotes M2 macrophage polarization to suppress atherosclerosis. Med. Sci. Monit. Basic Res. 23, 240–249 (2017).
Sas-Chen, A. et al. Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature 583, 638–643 (2020).
Yeste-Velasco, M., Linder, M. E. & Lu, Y. J. Protein -palmitoylation and cancer. Biochim. Biophys. Acta 1856, 107–120 (2015).
Lawlor, L. & Yang, X. B. Harnessing the HDAC-histone deacetylase enzymes, inhibitors and how these can be utilised in tissue engineering. Int. J. Oral. Sci. 11, 20 (2019).
Shen, Y., Wei, W. & Zhou, D. X. Histone acetylation enzymes coordinate metabolism and gene expression. Trends Plant Sci. 20, 614–621 (2015).
Wang, P., Wang, Z. & Liu, J. Role of HDACs in normal and malignant hematopoiesis. Mol. Cancer 19, 5 (2020).
Hu, T. et al. Metabolic rewiring by loss of Sirt5 promotes kras-induced pancreatic cancer progression. Gastroenterology 161, 1584–1600 (2021).
Sun, R. et al. Loss of SIRT5 promotes bile acid-induced immunosuppressive microenvironment and hepatocarcinogenesis. J. Hepatol. 77, 453–466 (2022).
Wu, S. et al. SIRT5 represses neurotrophic pathways and Abeta production in Alzheimer’s disease by targeting autophagy. ACS Chem. Neurosci. 12, 4428–4437 (2021).
Li, L. et al. SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat. Commun. 7, 12235 (2016).
Abmayr, S. M. & Workman, J. L. Histone lysine de-beta-hydroxybutyrylation by SIRT3. Cell Res. 29, 694–695 (2019).
Huang, H. et al. The regulatory enzymes and protein substrates for the lysine beta-hydroxybutyrylation pathway. Sci. Adv. 7, eabe2771 (2021).
Moreno-Yruela, C. et al. Class I histone deacetylases (HDAC1-3) are histone lysine delactylases. Sci. Adv. 8, eabi6696 (2022).
Bao, X. et al. Identification of ‘erasers’ for lysine crotonylated histone marks using a chemical proteomics approach. eLife 3, e02999 (2014).
Jiang, H. et al. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496, 110–113 (2013).
Burnaevskiy, N. et al. Proteolytic elimination of N-myristoyl modifications by the Shigella virulence factor IpaJ. Nature 496, 106–109 (2013).
Zhao, S., Zhang, X. & Li, H. Beyond histone acetylation-writing and erasing histone acylations. Curr. Opin. Struct. Biol. 53, 169–177 (2018).
Su, D. et al. Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106. Nature 483, 104–107 (2012).
Zhang, Y. et al. The ZZ domain of p300 mediates specificity of the adjacent HAT domain for histone H3. Nat. Struct. Mol. Biol. 25, 841–849 (2018).
Khan, A., Bridgers, J. B. & Strahl, B. D. Expanding the reader landscape of histone acylation. Structure 25, 571–573 (2017).
Li, Y. et al. Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Mol. Cell 62, 181–193 (2016).
Kabra, A. & Bushweller, J. The intrinsically disordered proteins MLLT3 (AF9) and MLLT1 (ENL) - multimodal transcriptional switches with roles in normal hematopoiesis, MLL fusion leukemia, and kidney cancer. J. Mol. Biol. 434, 167117 (2022).
Huang, B., Yang, X. D., Zhou, M. M., Ozato, K. & Chen, L. F. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol. Cell. Biol. 29, 1375–1387 (2009).
Roe, J. S., Mercan, F., Rivera, K., Pappin, D. J. & Vakoc, C. R. BET bromodomain inhibition suppresses the function of hematopoietic transcription factors in acute myeloid leukemia. Mol. Cell 58, 1028–1039 (2015).
Blee, A. M., Liu, S., Wang, L. & Huang, H. BET bromodomain-mediated interaction between ERG and BRD4 promotes prostate cancer cell invasion. Oncotarget 7, 38319–38332 (2016).
Shi, J. et al. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell 25, 210–225 (2014).
Qin, Z. Y. et al. BRD4 promotes gastric cancer progression and metastasis through acetylation-dependent stabilization of Snail. Cancer Res. 79, 4869–4881 (2019).
Lamonica, J. M. et al. Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes. Proc. Natl Acad. Sci. USA 108, E159–E168 (2011).
Cai, W. et al. PBRM1 acts as a p53 lysine-acetylation reader to suppress renal tumor growth. Nat. Commun. 10, 5800 (2019).
Hu, M., He, F., Thompson, E. W., Ostrikov, K. K. & Dai, X. Lysine acetylation, cancer hallmarks and emerging onco-therapeutic opportunities. Cancers 14, 346 (2022).
Shvedunova, M. & Akhtar, A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 23, 329–349 (2022).
Pietrocola, F., Galluzzi, L. Bravo-San Pedro, J. M., Madeo, F. & Kroemer, G. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 21, 805–821 (2015).
Weinert, B. T. et al. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol. Syst. Biol. 10, 716 (2014).
Ahmad, S. S. et al. Phosphoglycerate kinase 1 as a promoter of metastasis in colon cancer. Int. J. Oncol. 43, 586–590 (2013).
Duncan, L., Shay, C. & Teng, Y. PGK1: an essential player in modulating tumor metabolism. Methods Mol. Biol. 2343, 57–70 (2022).
Hu, H. et al. Acetylation of PGK1 promotes liver cancer cell proliferation and tumorigenesis. Hepatology 65, 515–528 (2017).
Liu, C. C. et al. ENO2 promotes cell proliferation, glycolysis, and glucocorticoid-resistance in acute lymphoblastic leukemia. Cell. Physiol. Biochem. 46, 1525–1535 (2018).
Carney, D. N. et al. Serum neuron-specific enolase: a marker for disease extent and response to therapy of small-cell lung cancer. Lancet 1, 583–585 (1982).
Zheng, Y. et al. Insulin-like growth factor 1-induced enolase 2 deacetylation by HDAC3 promotes metastasis of pancreatic cancer. Signal Transduct. Target. Ther. 5, 53 (2020).
Gu, L. et al. Amplification of glyceronephosphate O-acyltransferase and recruitment of USP30 stabilize DRP1 to promote hepatocarcinogenesis. Cancer Res. 78, 5808–5819 (2018).
Gu, L. et al. Stabilization of FASN by ACAT1-mediated GNPAT acetylation promotes lipid metabolism and hepatocarcinogenesis. Oncogene 39, 2437–2449 (2020).
Li, K. et al. TRIB3 promotes APL progression through stabilization of the oncoprotein PML-RAR alpha and inhibition of p53-mediated senescence. Cancer Cell 31, 697–710 (2017).
Shang, S. et al. TRIB3 reduces CD8(+) T cell infiltration and induces immune evasion by repressing the STAT1-CXCL10 axis in colorectal cancer. Sci. Transl. Med. 14, eabf0992 (2022).
Hua, F. et al. TRIB3 interacts with beta-catenin and TCF4 to increase stem cell features of colorectal cancer stem cells and tumorigenesis. Gastroenterology 156, 708–721 (2019).
Yu, J. J. et al. TRIB3-EGFR interaction promotes lung cancer progression and defines a therapeutic target. Nat. Commun. 11, 3660 (2020).
Yu, J. M. et al. TRIB3 supports breast cancer stemness by suppressing FOXO1 degradation and enhancing SOX2 transcription. Nat. Commun. 10, 5720 (2019).
Li, K. et al. TRIB3 promotes MYC-associated lymphoma development through suppression of UBE3B-mediated MYC degradation. Nat. Commun. 11, 6316 (2020).
Hua, F. et al. TRB3 links insulin/IGF to tumour promotion by interacting with p62 and impeding autophagic/proteasomal degradations. Nat. Commun. 6, 7951 (2015).
Hua, F. et al. TRB3 interacts with SMAD3 promoting tumor cell migration and invasion. J. Cell Sci. 124, 3235–3246 (2011).
Li, K. et al. Metformin suppresses melanoma progression by inhibiting KAT5-mediated SMAD3 acetylation, transcriptional activity and TRIB3 expression. Oncogene 37, 2967–2981 (2018).
Mc Gee, M. M. Targeting the mitotic catastrophe signaling pathway in cancer. Mediators Inflamm. 2015, 146282 (2015).
Vitale, I., Galluzzi, L., Castedo, M. & Kroemer, G. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat. Rev. Mol. Cell Biol. 12, 385–392 (2011).
Yi, F. et al. The deacetylation-phosphorylation regulation of SIRT2-SMC1A axis as a mechanism of antimitotic catastrophe in early tumorigenesis. Sci. Adv. 7, eabe5518 (2021).
Rios Garcia, M. et al. Acetyl-CoA carboxylase 1-dependent protein acetylation controls breast cancer metastasis and recurrence. Cell Metab. 26, 842–855 (2017).
Kryczek, I. et al. IL-22(+)CD4(+) T cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L. Immunity 40, 772–784 (2014).
Bourguignon, L. Y., Wong, G. & Shiina, M. Up-regulation of histone methyltransferase, DOT1L, by matrix hyaluronan promotes microRNA-10 expression leading to tumor cell invasion and chemoresistance in cancer stem cells from head and neck squamous cell carcinoma. J. Biol. Chem. 291, 10571–10585 (2016).
Liu, C. et al. CBP mediated DOT1L acetylation confers DOT1L stability and promotes cancer metastasis. Theranostics 10, 1758–1776 (2020).
Zhang, W., Zhang, H., Ning, L., Li, B. & Bao, M. Quantitative proteomic analysis provides novel insights into cold stress responses in Petunia seedlings. Front. Plant Sci. 7, 136 (2016).
Dong, Z., Yang, L. & Lai, D. KLF5 strengthens drug resistance of ovarian cancer stem-like cells by regulating survivin expression. Cell Prolif. 46, 425–435 (2013).
Zhang, B. et al. Acetylation of KLF5 maintains EMT and tumorigenicity to cause chemoresistant bone metastasis in prostate cancer. Nat. Commun. 12, 1714 (2021).
Kong, Y. et al. Histone deacetylase inhibitors (HDACi) promote KLF5 ubiquitination and degradation in basal-like breast cancer. Int. J. Biol. Sci. 18, 2104–2115 (2022).
Liu, Y., Tavana, O. & Gu, W. p53 modifications: exquisite decorations of the powerful guardian. J. Mol. Cell Biol. 11, 564–577 (2019).
Reed, S. M. & Quelle, D. E. P53 acetylation: regulation and consequences. Cancers 7, 30–69 (2014).
Wang, S. J. et al. Acetylation is crucial for p53-mediated ferroptosis and tumor suppression. Cell Rep. 17, 366–373 (2016).
Rodriguez, M. S., Desterro, J. M., Lain, S., Lane, D. P. & Hay, R. T. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol. Cell. Biol. 20, 8458–8467 (2000).
Liu, L. et al. P53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 19, 1202–120 (1999).
Blank, C. & Mackensen, A. Contribution of the PD-L1/PD-1 pathway to T-cell exhaustion: an update on implications for chronic infections and tumor evasion. Cancer Immunol. Immunother. 56, 739–745 (2007).
Ai, L., Xu, A. & Xu, J. Roles of PD-1/PD-L1 pathway: signaling, cancer, and beyond. Adv. Exp. Med. Biol. 1248, 33–59 (2020).
Gaikwad, S., Agrawal, M. Y., Kaushik, I., Ramachandran, S. & Srivastava, S. K. Immune checkpoint proteins: signaling mechanisms and molecular interactions in cancer immunotherapy. Semin. Cancer Biol. 86, 137–150 (2022).
Gao, Y. et al. Acetylation-dependent regulation of PD-L1 nuclear translocation dictates the efficacy of anti-PD-1 immunotherapy. Nat. Cell Biol. 22, 1064–1075 (2020).
Ma, L. et al. Overexpression of the transcription factor MEF2D in hepatocellular carcinoma sustains malignant character by suppressing G2-M transition genes. Cancer Res. 74, 1452–1462 (2014).
Xiang, J. et al. Disruption of SIRT7 increases the efficacy of checkpoint inhibitor via MEF2D regulation of programmed cell death 1 ligand 1 in hepatocellular carcinoma cells. Gastroenterology 158, 664–678 (2020).
Ma, K., Chan, J. K., Zhu, G. & Wu, Z. Myocyte enhancer factor 2 acetylation by p300 enhances its DNA binding activity, transcriptional activity, and myogenic differentiation. Mol. Cell. Biol. 25, 3575–3582 (2005).
Prajapati, K., Perez, C., Rojas, L. B. P., CHke, B. & Guevara-Patino, J. A. Functions of NKG2D in CD8(+) T cells: an opportunity for immunotherapy. Cell. Mol. Immunol. 15, 470–479 (2018).
Waldhauer, I. & Steinle, A. Proteolytic release of soluble UL16-binding protein 2 from tumor cells. Cancer Res. 66, 2520–2526 (2006).
Ashiru, O. et al. Natural killer cell cytotoxicity is suppressed by exposure to the human NKG2D ligand MICA*008 that is shed by tumor cells in exosomes. Cancer Res. 70, 481–489 (2010).
Hu, J. et al. Induction of NKG2D ligands on solid tumors requires tumor-specific CD8(+) T Cells and histone acetyltransferases. Cancer Immunol. Res. 5, 300–311 (2017).
Hu, J., Xia, X., Zhao, Q. & Li, S. Lysine acetylation of NKG2D ligand Rae-1 stabilizes the protein and sensitizes tumor cells to NKG2D immune surveillance. Cancer Lett. 502, 143–153 (2021).
Fang, H. S. et al. SIRT1 induces the accumulation of TAMs at colorectal cancer tumor sites via the CXCR4/CXCL12 axis. Cell. Immunol. 371, 104458 (2022).
Zhou, Y. et al. E3 ubiquitin ligase SIAH1 mediates ubiquitination and degradation of TRB3. Cell. Signal. 20, 942–948 (2008).
Yu, B. et al. KAT6A acetylation of SMAD3 regulates myeloid-derived suppressor cell recruitment, metastasis, and immunotherapy in triple-negative breast cancer. Adv. Sci. 9, e2105793 (2022).
Yu, T. et al. Modulation of M2 macrophage polarization by the crosstalk between Stat6 and Trim24. Nat. Commun. 10, 4353 (2019).
Caballero, B. et al. Interplay of pathogenic forms of human tau with different autophagic pathways. Aging Cell 17, e12692 (2018).
Caballero, B. et al. Acetylated tau inhibits chaperone-mediated autophagy and promotes tau pathology propagation in mice. Nat. Commun. 12, 2238 (2021).
Alquezar, C. et al. TSC1 loss increases risk for tauopathy by inducing tau acetylation and preventing tau clearance via chaperone-mediated autophagy. Sci. Adv. 7, eabg3897 (2021).
Trzeciakiewicz, H. et al. An HDAC6-dependent surveillance mechanism suppresses tau-mediated neurodegeneration and cognitive decline. Nat. Commun. 11, 5522 (2020).
Choi, H. et al. Acetylation changes tau interactome to degrade tau in Alzheimer’s disease animal and organoid models. Aging Cell 19, e13081 (2020).
Zhang, Y. et al. SIRT2-mediated deacetylation and deubiquitination of C/EBPbeta prevents ethanol-induced liver injury. Cell Discov. 7, 93 (2021).
Ren, H. et al. Sirtuin 2 prevents liver steatosis and metabolic disorders by deacetylation of hepatocyte nuclear factor 4alpha. Hepatology 74, 723–740 (2021).
Wang, T. et al. Acetylation of lactate dehydrogenase B drives NAFLD progression by impairing lactate clearance. J. Hepatol. 74, 1038–1052 (2021).
He, M. et al. An acetylation switch of the NLRP3 inflammasome regulates aging-associated chronic inflammation and insulin resistance. Cell Metab. 31, 580–591 (2020).
Song, Z. M. et al. KAT5 acetylates cGAS to promote innate immune response to DNA virus. Proc. Natl Acad. Sci. USA 117, 21568–21575 (2020).
Zhang, Z. et al. Acetylation-dependent deubiquitinase OTUD3 controls MAVS activation in innate antiviral immunity. Mol. Cell 79, 304–319 (2020).
Zhu, N. et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020).
Park, H. H. et al. Acetylated K676 TGFBIp as a severity diagnostic blood biomarker for SARS-CoV-2 pneumonia. Sci. Adv. 6, eabc1564 (2020).
Choudhary, C., Weinert, B. T., Nishida, Y., Verdin, E. & Mann, M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 15, 536–550 (2014).
O’Shea, E. K., Klemm, J. D., Kim, P. S. & Alber, T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254, 539–544 (1991).
Weinert, B. T. et al. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 4, 842–851 (2013).
Gut, P. et al. SUCLA2 mutations cause global protein succinylation contributing to the pathomechanism of a hereditary mitochondrial disease. Nat. Commun. 11, 5927 (2020).
Li, X. et al. Lysine-222 succinylation reduces lysosomal degradation of lactate dehydrogenase a and is increased in gastric cancer. J. Exp. Clin. Cancer Res. 39, 172 (2020).
Yang, X. et al. SHMT2 desuccinylation by SIRT5 drives cancer cell proliferation. Cancer Res. 78, 372–386 (2018).
Wang, C. et al. CPT1A-mediated succinylation of S100A10 increases human gastric cancer invasion. J. Cell. Mol. Med. 23, 293–305 (2019).
Guo, Z. et al. Systematic Proteome and lysine succinylome analysis reveals enhanced cell migration by hyposuccinylation in esophageal squamous cell carcinoma. Mol. Cell. Proteom. 20, 100053 (2021).
Lu, W. et al. Succinylation regulators promote clear cell renal cell carcinoma by immune regulation and RNA N6-methyladenosine methylation. Front. Cell Dev. Biol. 9, 622198 (2021).
Tannahill, G. M. et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature 496, 238–242 (2013).
Nair, S. et al. Lipopolysaccharide-induced alteration of mitochondrial morphology induces a metabolic shift in microglia modulating the inflammatory response in vitro and in vivo. Glia 67, 1047–1061 (2019).
Palsson-McDermott, E. M. et al. Pyruvate kinase M2 regulates hif-1alpha activity and IL-1beta induction and is a critical determinant of the Warburg effect in LPS-activated macrophages. Cell Metab. 21, 65–80 (2015).
Wang, F. et al. SIRT5 desuccinylates and activates pyruvate kinase M2 to block macrophage IL-1beta production and to prevent DSS-induced colitis in mice. Cell Rep. 19, 2331–2344 (2017).
Liu, Q. et al. The global succinylation of SARS-CoV-2-infected host cells reveals drug targets. Proc. Natl Acad. Sci. USA 119, e2123065119 (2022).
Ali, H. R. et al. Defining decreased protein succinylation of failing human cardiac myofibrils in ischemic cardiomyopathy. J. Mol. Cell. Cardiol. 138, 304–317 (2020).
Zhang, Y. et al. Sirt5-mediated desuccinylation of OPTN protects retinal ganglion cells from autophagic flux blockade in diabetic retinopathy. Cell Death Discov. 8, 63 (2022).
Cheng, Y., Hou, T., Ping, J., Chen, G. & Chen, J. Quantitative succinylome analysis in the liver of non-alcoholic fatty liver disease rat model. Proteome Sci. 14, 3 (2016).
Cheung, N., Mitchell, P. & Wong, T. Y. Diabetic retinopathy. Lancet 376, 124–136 (2010).
Guo, Y. et al. NF- kappa B/HDAC1/SREBP1c pathway mediates the inflammation signal in progression of hepatic steatosis. Acta Pharm. Sin. B 10, 825–836 (2020).
Yang, Y. et al. Altered succinylation of mitochondrial proteins, APP and tau in Alzheimer’s disease. Nat. Commun. 13, 159 (2022).
Masters, C. L. et al. Alzheimer’s disease. Nat. Rev. Dis. Prim. 1, 15056 (2015).
Matrone, C., Iannuzzi, F. & Annunziato, L. The Y682ENPTY687 motif of APP: progress and insights toward a targeted therapy for Alzheimer’s disease patients. Ageing Res. Rev. 52, 120–128 (2019).
Ballard, C. et al. Alzheimer’s disease. Lancet 377, 1019–1031 (2011).
He, Z. et al. Amyloid-beta plaques enhance Alzheimer’s brain tau-seeded pathologies by facilitating neuritic plaque tau aggregation. Nat. Med. 24, 29–38 (2018).
Peng, C. et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell. Proteom. 10, M111 012658 (2011).
Bowman, C. E. et al. The mammalian malonyl-CoA synthetase ACSF3 is required for mitochondrial protein malonylation and metabolic efficiency. Cell Chem. Biol. 24, 673–684 (2017).
Folmes, C. D. & Lopaschuk, G. D. Role of malonyl-CoA in heart disease and the hypothalamic control of obesity. Cardiovasc. Res. 73, 278–287 (2007).
Zhao, Z. et al. Rosiglitazone and fenofibrate improve insulin sensitivity of pre-diabetic OLETF rats by reducing malonyl-CoA levels in the liver and skeletal muscle. Life Sci. 84, 688–695 (2009).
Bandyopadhyay, G. K., Yu, J. G., Ofrecio, J. & Olefsky, J. M. Increased malonyl-CoA levels in muscle from obese and type 2 diabetic subjects lead to decreased fatty acid oxidation and increased lipogenesis; thiazolidinedione treatment reverses these defects. Diabetes 55, 2277–2285 (2006).
Prentki, M., Joly, E., El-Assaad, W. & Roduit, R. Malonyl-CoA signaling, lipid partitioning, and glucolipotoxicity: role in beta-cell adaptation and failure in the etiology of diabetes. Diabetes 51, S405–S413 (2002).
Rardin, M. J. et al. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 18, 920–933 (2013).
Du, Y. et al. Lysine malonylation is elevated in type 2 diabetic mouse models and enriched in metabolic associated proteins. Mol. Cell. Proteom. 14, 227–236 (2015).
Bruning, U. et al. Impairment of angiogenesis by fatty acid synthase inhibition involves mTOR malonylation. Cell Metab. 28, 866–880 (2018).
Pocaterra, A. et al. F-actin dynamics regulates mammalian organ growth and cell fate maintenance. J. Hepatol. 71, 130–142 (2019).
Plessner, M., Melak, M., Chinchilla, P., Baarlink, C. & Grosse, R. Nuclear F-actin formation and reorganization upon cell spreading. J. Biol. Chem. 290, 11209–11216 (2015).
Huang, Q. et al. TFAM loss induces nuclear actin assembly upon mDia2 malonylation to promote liver cancer metastasis. EMBO J. 41, e110324 (2022).
Baarlink, C. et al. A transient pool of nuclear F-actin at mitotic exit controls chromatin organization. Nat. Cell Biol. 19, 1389–1399 (2017).
Rodriguez-Prados, J. C. et al. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J. Immunol. 185, 605–614 (2010).
Galvan-Pena, S. et al. Malonylation of GAPDH is an inflammatory signal in macrophages. Nat. Commun. 10, 338 (2019).
Jha, A. K. et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–430 (2015).
Peoples, J. N. et al. Loss of the mitochondrial phosphate carrier SLC25A3 induces remodeling of the cardiac mitochondrial protein acylome. Am. J. Physiol. Cell Physiol. 321, C519–C534 (2021).
Wu, L. F. et al. Global profiling of protein lysine malonylation in mouse cardiac hypertrophy. J. Proteom. 266, 104667 (2022).
Du, Y. et al. SIRT5 deacylates metabolism-related proteins and attenuates hepatic steatosis in ob/ob mice. EBioMedicine 36, 347–357 (2018).
Jiang, G., Li, C., Lu, M., Lu, K. & Li, H. Protein lysine crotonylation: past, present, perspective. Cell Death Dis. 12, 703 (2021).
Xu, W. et al. Global profiling of crotonylation on non-histone proteins. Cell Res. 27, 946–949 (2017).
Wei, W. et al. Large-scale identification of protein crotonylation reveals its role in multiple cellular functions. J. Proteome Res. 16, 1743–1752 (2017).
Chen, Y. Z. et al. NhKcr: a new bioinformatics tool for predicting crotonylation sites on human nonhistone proteins based on deep learning. Brief. Bioinform. 22, bbab146 (2021).
Dou, L., Zhang, Z., Xu, L. & Zou, Q. IKcr_CNN: a novel computational tool for imbalance classification of human nonhistone crotonylation sites based on convolutional neural networks with focal loss. Comput. Struct. Biotechnol. J. 20, 3268–3279 (2022).
Tang, X. et al. Short-chain enoyl-CoA hydratase mediates histone crotonylation and contributes to cardiac homeostasis. Circulation 143, 1066–1069 (2021).
Cai, W. et al. Modulating lysine crotonylation in cardiomyocytes improves myocardial outcomes. Circ. Res. 131, 456–472 (2022).
Wan, J., Liu, H. & Ming, L. Lysine crotonylation is involved in hepatocellular carcinoma progression. Biomed. Pharmacother. 111, 976–982 (2019).
Zhan, P. et al. Alpha-enolase promotes tumorigenesis and metastasis via regulating AMPK/mTOR pathway in colorectal cancer. Mol. Carcinog. 56, 1427–1437 (2017).
Yuan, Z. et al. Colorectal cancer cell intrinsic fibroblast activation protein alpha binds to Enolase1 and activates NF-kappaB pathway to promote metastasis. Cell Death Dis. 12, 543 (2021).
Hou, J. Y. et al. Upregulation of alpha enolase (ENO1) crotonylation in colorectal cancer and its promoting effect on cancer cell metastasis. Biochem. Biophys. Res. Commun. 578, 77–83 (2021).
Yu, C. et al. Oral squamous cancer cell exploits hnRNP A1 to regulate cell cycle and proliferation. J. Cell. Physiol. 230, 2252–2261 (2015).
Zhang, H. et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol. Cancer 19, 43 (2020).
Jiang, R. et al. Esculetin inhibits endometrial cancer proliferation and promotes apoptosis via hnRNPA1 to downregulate BCLXL and XIAP. Cancer Lett. 521, 308–321 (2021).
Han, X. et al. P300-catalyzed lysine crotonylation promotes the proliferation, invasion, and migration of HeLa cells via heterogeneous nuclear ribonucleoprotein A1. Anal. Cell. Pathol. 2020, 5632342 (2020).
Kolb, H. et al. Ketone bodies: from enemy to friend and guardian angel. BMC Med. 19, 313 (2021).
Zhang, H. et al. MTA2 triggered R-loop trans-regulates BDH1-mediated beta-hydroxybutyrylation and potentiates propagation of hepatocellular carcinoma stem cells. Signal Transduct. Target. Ther. 6, 135 (2021).
Chen, L., Miao, Z. & Xu, X. Beta-hydroxybutyrate alleviates depressive behaviors in mice possibly by increasing the histone3-lysine9-beta-hydroxybutyrylation. Biochem. Biophys. Res. Commun. 490, 117–122 (2017).
Dmitrieva-Posocco, O. et al. Beta-hydroxybutyrate suppresses colorectal cancer. Nature 605, 160–165 (2022).
Liu, K. et al. P53 beta-hydroxybutyrylation attenuates p53 activity. Cell Death Dis. 10, 243 (2019).
Shukla, S. K. et al. Metabolic reprogramming induced by ketone bodies diminishes pancreatic cancer cachexia. Cancer Metab. 2, 18 (2014).
Kang, H. B. et al. Metabolic rewiring by oncogenic BRAF V600E links ketogenesis pathway to BRAF-MEK1 signaling. Mol. Cell 59, 345–358 (2015).
Doherty, J. R. & Cleveland, J. L. Targeting lactate metabolism for cancer therapeutics. J. Clin. Invest. 123, 3685–3692 (2013).
Jones, A. E. et al. Lactate clearance vs central venous oxygen saturation as goals of early sepsis therapy: a randomized clinical trial. J. Am. Med. Assoc. 303, 739–746 (2010).
de la Cruz-Lopez, K. G., Castro-Munoz, L. J., Reyes-Hernandez, D. O., Garcia-Carranca, A. & Manzo-Merino, J. Lactate in the regulation of tumor microenvironment and therapeutic approaches. Front. Oncol. 9, 1143 (2019).
Watson, M. J. et al. Metabolic support of tumour-infiltrating regulatory T cells by lactic acid. Nature 591, 645–651 (2021).
Apicella, M. et al. Increased lactate secretion by cancer cells sustains non-cell-autonomous adaptive resistance to MET and EGFR targeted therapies. Cell Metab. 28, 848–865 (2018).
Tasdogan, A. et al. Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature 577, 115–120 (2020).
Shu, E., Farshidpour, L., Young, M., Darracq, M. & Ives Tallman, C. Utility of point-of-care synovial lactate to identify septic arthritis in the emergency department. Am. J. Emerg. Med. 37, 502–505 (2019).
Pan, L. et al. Demethylzeylasteral targets lactate by inhibiting histone lactylation to suppress the tumorigenicity of liver cancer stem cells. Pharmacol. Res. 181, 106270 (2022).
Yang, J. et al. A positive feedback loop between inactive VHL-triggered histone lactylation and PDGFRbeta signaling drives clear cell renal cell carcinoma progression. Int. J. Biol. Sci. 18, 3470–3483 (2022).
Xiong, J. et al. Lactylation-driven METTL3-mediated RNA m(6)A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol. Cell 82, 1660–1677 (2022).
Cui, H. et al. Lung myofibroblasts promote macrophage profibrotic activity through lactate-induced histone lactylation. Am. J. Respir. Cell Mol. Biol. 64, 115–125 (2021).
Jiang, J. et al. Lactate modulates cellular metabolism through histone lactylation-mediated gene expression in non-small cell lung cancer. Front. Oncol. 11, 647559 (2021).
Pan, R. Y. et al. Positive feedback regulation of microglial glucose metabolism by histone H4 lysine 12 lactylation in Alzheimer’s disease. Cell Metab. 34, 634–648 (2022).
Wan, N. et al. Cyclic immonium ion of lactyllysine reveals widespread lactylation in the human proteome. Nat. Methods 19, 854–864 (2022).
Sun, Y., Chen, Y. & Peng, T. A bioorthogonal chemical reporter for the detection and identification of protein lactylation. Chem. Sci. 13, 6019–6027 (2022).
Yang, D. et al. Identification of lysine-lactylated substrates in gastric cancer cells. iScience 25, 104630 (2022).
Gu, J. et al. Tumor metabolite lactate promotes tumorigenesis by modulating MOESIN lactylation and enhancing TGF-beta signaling in regulatory T cells. Cell Rep. 39, 110986 (2022).
Correa, T. D. et al. Time course of blood lactate levels, inflammation, and mitochondrial function in experimental sepsis. Crit. Care 21, 105 (2017).
Deng, M. et al. The endotoxin delivery protein HMGB1 mediates caspase-11-dependent lethality in Sepsis. Immunity 49, 740–753 (2018).
Fhu, C. W. & Ali, A. Protein lipidation by palmitoylation and myristoylation in cancer. Front. Cell Dev. Biol. 9, 673647 (2021).
Xiong, W. H., Qin, M. & Zhong, H. Myristoylation alone is sufficient for PKA catalytic subunits to associate with the plasma membrane to regulate neuronal functions. Proc. Natl Acad. Sci. USA 118, e2021658118 (2021).
Herzig, S. & Shaw, R. J. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 19, 121–135 (2018).
Bouchier, I. A. Some experiences of ships’ surgeons during the early days of the sperm whale fishery. Br. Med. J. 285, 1811–1813 (1982).
Lu, X. et al. AMPK protects against alcohol-induced liver injury through UQCRC2 to up-regulate mitophagy. Autophagy 17, 3622–3643 (2021).
Liang, J. et al. Myristoylation confers noncanonical AMPK functions in autophagy selectivity and mitochondrial surveillance. Nat. Commun. 6, 7926 (2015).
Sun, Y. et al. N-myristoyltransferase-1 deficiency blocks myristoylation of LAMTOR1 and inhibits bladder cancer progression. Cancer Lett. 529, 126–138 (2022).
Chen, Y. C. et al. N-myristoyltransferase-1 is necessary for lysosomal degradation and mTORC1 activation in cancer cells. Sci. Rep. 10, 11952 (2020).
Kim, S. et al. Blocking myristoylation of Src inhibits its kinase activity and suppresses prostate cancer progression. Cancer Res. 77, 6950–6962 (2017).
Li, Q. et al. Pharmacologically targeting the myristoylation of the scaffold protein FRS2alpha inhibits FGF/FGFR-mediated oncogenic signaling and tumor progression. J. Biol. Chem. 293, 6434–6448 (2018).
Dieci, M. V., Arnedos, M., Andre, F. & Soria, J. C. Fibroblast growth factor receptor inhibitors as a cancer treatment: from a biologic rationale to medical perspectives. Cancer Discov. 3, 264–279 (2013).
Subbiah, V. et al. FIGHT-101, a first-in-human study of potent and selective FGFR 1-3 inhibitor pemigatinib in pan-cancer patients with FGF/FGFR alterations and advanced malignancies. Ann. Oncol. 33, 522–533 (2022).
Bohrer, L. R. et al. Activation of the FGFR-STAT3 pathway in breast cancer cells induces a hyaluronan-rich microenvironment that licenses tumor formation. Cancer Res. 74, 374–386 (2014).
Elstrom, R. L. et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 64, 3892–3899 (2004).
Adam, R. M. et al. Cholesterol sensitivity of endogenous and myristoylated Akt. Cancer Res. 67, 6238–6246 (2007).
Zhang, Q. Y. et al. Metabolic reprogramming of ovarian cancer involves ACSL1-mediated metastasis stimulation through upregulated protein myristoylation. Oncogene 40, 97–111 (2021).
Zhu, G. Q. et al. N-myristoylation by NMT1 is POTEE-dependent to stimulate liver tumorigenesis via differentially regulating ubiquitination of targets. Front. Oncol. 11, 681366 (2021).
Wen, Z. et al. N-myristoyltransferase deficiency impairs activation of kinase AMPK and promotes synovial tissue inflammation. Nat. Immunol. 20, 313–325 (2019).
Wang, B. et al. Protein N-myristoylation: functions and mechanisms in control of innate immunity. Cell. Mol. Immunol. 18, 878–888 (2021).
Resh, M. D. A myristoyl switch regulates membrane binding of HIV-1 Gag. Proc. Natl Acad. Sci. USA 101, 417–418 (2004).
Mousnier, A. et al. Fragment-derived inhibitors of human N-myristoyltransferase block capsid assembly and replication of the common cold virus. Nat. Chem. 10, 599–606 (2018).
Lindwasser, O. W. & Resh, M. D. Myristoylation as a target for inhibiting HIV assembly: unsaturated fatty acids block viral budding. Proc. Natl Acad. Sci. USA 99, 13037–13042 (2002).
Ji, Y., Wang, D., Zhang, B. & Lu, H. Bergenin mmeliorates MPTP-induced Parkinson’s disease by activating PI3K/Akt signaling pathway. J. Alzheimers Dis. 72, 823–833 (2019).
Huang, Y. et al. Neuroprotection against Parkinson’s disease through the activation of Akt/GSK3beta signaling pathway by Tovophyllin A. Front. Neurosci. 14, 723 (2020).
Ries, V. et al. Oncoprotein Akt/PKB induces trophic effects in murine models of Parkinson’s disease. Proc. Natl Acad. Sci. USA 103, 18757–18762 (2006).
Pei, Z., Xiao, Y., Meng, J., Hudmon, A. & Cummins, T. R. Cardiac sodium channel palmitoylation regulates channel availability and myocyte excitability with implications for arrhythmia generation. Nat. Commun. 7, 12035 (2016).
Boucher, J. M. et al. Dynamic alterations in decoy VEGF receptor-1 stability regulate angiogenesis. Nat. Commun. 8, 15699 (2017).
Main, A., Robertson-Gray, O. & Fuller, W. Cyclophilin D palmitoylation and permeability transition: a new twist in the tale of myocardial ischaemia-reperfusion injury. Cardiovasc. Res. 117, 15–17 (2021).
Wei, X. et al. Endothelial palmitoylation cycling coordinates vessel remodeling in peripheral artery disease. Circ. Res. 127, 249–265 (2020).
Wei, X. et al. Fatty acid synthase modulates intestinal barrier function through palmitoylation of mucin 2. Cell Host Microbe 11, 140–152 (2012).
Zhang, M. et al. A STAT3 palmitoylation cycle promotes TH17 differentiation and colitis. Nature 586, 434–439 (2020).
Zhou, L. et al. Palmitoylation restricts SQSTM1/p62-mediated autophagic degradation of NOD2 to modulate inflammation. Cell Death Differ. 29, 1541–1551 (2022).
Hansen, A. L. et al. Nitro-fatty acids are formed in response to virus infection and are potent inhibitors of STING palmitoylation and signaling. Proc. Natl Acad. Sci. USA 115, E7768–E7775 (2018).
Adekola, K., Rosen, S. T. & Shanmugam, M. Glucose transporters in cancer metabolism. Curr. Opin. Oncol. 24, 650–654 (2012).
Amann, T. et al. GLUT1 expression is increased in hepatocellular carcinoma and promotes tumorigenesis. Am. J. Pathol. 174, 1544–1552 (2009).
Wu, Q. et al. GLUT1 inhibition blocks growth of RB1-positive triple negative breast cancer. Nat. Commun. 11, 420 (2020).
Ancey, P. B., Contat, C. & Meylan, E. Glucose transporters in cancer—from tumor cells to the tumor microenvironment. FEBS J. 285, 2926–2943 (2018).
Zhang, Z. et al. DHHC9-mediated GLUT1 S-palmitoylation promotes glioblastoma glycolysis and tumorigenesis. Nat. Commun. 12, 5872 (2021).
Balsa, E. et al. ER and nutrient stress promote assembly of respiratory chain supercomplexes through the PERK-eIF2alpha axis. Mol. Cell 74, 877–890 (2019).
Shan, B. et al. The metabolic ER stress sensor IRE1alpha suppresses alternative activation of macrophages and impairs energy expenditure in obesity. Nat. Immunol. 18, 519–529 (2017).
Jin, J. K. et al. ATF6 decreases myocardial ischemia/reperfusion damage and links ER stress and oxidative stress signaling pathways in the heart. Circ. Res. 120, 862–875 (2017).
Chen, X. et al. Protein palmitoylation regulates cell survival by modulating XBP1 activity in glioblastoma multiforme. Mol. Ther. Oncolytics 17, 518–530 (2020).
Lipskaia, L. & Lompre, A. M. Alteration in temporal kinetics of Ca2+ signaling and control of growth and proliferation. Biol. Cell 96, 55–68 (2004).
Chen, P. et al. Erianin, a novel dibenzyl compound in Dendrobium extract, inhibits lung cancer cell growth and migration via calcium/calmodulin-dependent ferroptosis. Signal Transduct. Target. Ther. 5, 51 (2020).
Monteith, G. R., McAndrew, D., Faddy, H. M. & Roberts-Thomson, S. J. Calcium and cancer: targeting Ca2+ transport. Nat. Rev. Cancer 7, 519–530 (2007).
Fredericks, G. J. et al. Stable expression and function of the inositol 1,4,5-triphosphate receptor requires palmitoylation by a DHHC6/selenoprotein K complex. Proc. Natl Acad. Sci. USA 111, 16478–16483 (2014).
Parekh, A. B. & Putney, J. W. Jr Store-operated calcium channels. Physiol. Rev. 85, 757–810 (2005).
Marciel, M. P. et al. Selenoprotein K deficiency inhibits melanoma by reducing calcium flux required for tumor growth and metastasis. Oncotarget 9, 13407–13422 (2018).
Huang, Y. H. et al. Claudin-3 gene silencing with siRNA suppresses ovarian tumor growth and metastasis. Proc. Natl Acad. Sci. USA 106, 3426–3430 (2009).
Yuan, M. et al. ZDHHC12-mediated claudin-3 S-palmitoylation determines ovarian cancer progression. Acta Pharm. Sin. B 10, 1426–1439 (2020).
Lynch, S. J. et al. The differential palmitoylation states of N-Ras and H-Ras determine their distinct golgi subcompartment localizations. J. Cell. Physiol. 230, 610–619 (2015).
Cuiffo, B. & Ren, R. Palmitoylation of oncogenic NRAS is essential for leukemogenesis. Blood 115, 3598–3605 (2010).
Liu, P. et al. Palmitoylacyltransferase Zdhhc9 inactivation mitigates leukemogenic potential of oncogenic Nras. Leukemia 30, 1225–1228 (2016).
Kadry, Y. A., Lee, J. Y. & Witze, E. S. Regulation of EGFR signalling by palmitoylation and its role in tumorigenesis. Open Biol. 11, 210033 (2021).
Raimondi, S. et al. MC1R variants, melanoma and red hair color phenotype: a meta-analysis. Int. J. Cancer 122, 2753–2760 (2008).
Abdel-Mallek, Z. A., Knittel, J., Kadekaro, A. L., Swope, V. B. & Starner, R. The melanocortin 1 receptor and the UV response of human melanocytes—a shift in paradigm. Photochem. Photobiol. 84, 501–508 (2008).
Chen, S. et al. Palmitoylation-dependent activation of MC1R prevents melanomagenesis. Nature 549, 399–403 (2017).
Lin, H. Protein cysteine palmitoylation in immunity and inflammation. FEBS J. 288, 7043–7059 (2021).
Yang, Y. et al. Palmitoylation stabilizes PD-L1 to promote breast tumor growth. Cell Res. 29, 83–86 (2019).
Yao, H. et al. Inhibiting PD-L1 palmitoylation enhances T-cell immune responses against tumours. Nat. Biomed. Eng. 3, 306–317 (2019).
Yao, H. et al. A peptidic inhibitor for PD-1 palmitoylation targets its expression and functions. RSC Chem. Biol. 2, 192–205 (2021).
Gao, J. et al. Loss of IFN-gamma pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 167, 397–404 (2016).
Du, W. et al. Loss of optineurin drives cancer immune evasion via palmitoylation-dependent IFNGR1 lysosomal sorting and degradation. Cancer Discov. 11, 1826–1843 (2021).
Li, D., Liu, Y., Lu, Y., Gao, S. & Zhang, L. Palmitoylation of SARS-CoV-2 S protein is critical for S-mediated syncytia formation and virus entry. J. Med. Virol. 94, 342–348 (2022).
Xie, F. et al. Engineering extracellular vesicles enriched with palmitoylated ACE2 as COVID-19 therapy. Adv. Mater. 33, e2103471 (2021).
Andrew, R. J. et al. Lack of BACE1 S-palmitoylation reduces amyloid burden and mitigates memory deficits in transgenic mouse models of Alzheimer’s disease. Proc. Natl Acad. Sci. USA 114, E9665–E9674 (2017).
Ross, C. A. & Tabrizi, S. J. Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 10, 83–98 (2011).
Singaraja, R. R. et al. Altered palmitoylation and neuropathological deficits in mice lacking HIP14. Hum. Mol. Genet. 20, 3899–3909 (2011).
Yanai, A. et al. Palmitoylation of huntingtin by HIP14 is essential for its trafficking and function. Nat. Neurosci. 9, 824–831 (2006).
Virlogeux, A. et al. Increasing brain palmitoylation rescues behavior and neuropathology in Huntington disease mice. Sci. Adv. 7, eabb0799 (2021).
Liu, L. et al. Astrocyte secretes IL-6 to modulate PSD-95 palmitoylation in basolateral amygdala and depression-like behaviors induced by peripheral nerve injury. Brain Behav. Immun. 104, 139–154 (2022).
Gorinski, N. et al. Attenuated palmitoylation of serotonin receptor 5-HT1A affects receptor function and contributes to depression-like behaviors. Nat. Commun. 10, 3924 (2019).
Zhao, J. et al. Glucose-sensitive acetylation of Seryl tRNA synthetase regulates lipid synthesis in breast cancer. Signal Transduct. Target. Ther. 6, 303 (2021).
Elumalai, S., Karunakaran, U., Moon, J. S. & Won, K. C. High glucose-induced PRDX3 acetylation contributes to glucotoxicity in pancreatic beta-cells: prevention by teneligliptin. Free Radic. Biol. Med. 160, 618–629 (2020).
Du, J. et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334, 806–809 (2011).
Imai, S., Armstrong, C. M., Kaeberlein, M. & Guarente, L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 (2000).
Obradovic, M. et al. Leptin and obesity: role and clinical implication. Front. Endocrinol. 12, 585887 (2021).
Shimazu, T. et al. Suppression of oxidative stress by beta-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science 339, 211–214 (2013).
Hong, S. H., Kang, M., Lee, K. S. & Yu, K. High fat diet-induced TGF-beta/Gbb signaling provokes insulin resistance through the tribbles expression. Sci. Rep. 6, 30265 (2016).
Brandon, E. L. et al. Obesity promotes melanoma tumor growth: role of leptin. Cancer Biol. Ther. 8, 1871–1879 (2009).
Schwer, B. et al. Calorie restriction alters mitochondrial protein acetylation. Aging Cell 8, 604–606 (2009).
Huang, H. et al. L-carnitine is an endogenous HDAC inhibitor selectively inhibiting cancer cell growth in vivo and in vitro. PLoS ONE 7, e49062 (2012).
Hait, N. C. et al. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science 325, 1254–1257 (2009).
Watson, P. J. et al. Insights into the activation mechanism of class I HDAC complexes by inositol phosphates. Nat. Commun. 7, 11262 (2016).
Le Cam, L. et al. E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell 127, 775–788 (2006).
Kramer, O. H. et al. A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev. 23, 223–235 (2009).
Ivanov, G. S. et al. Methylation-acetylation interplay activates p53 in response to DNA damage. Mol. Cell. Biol. 27, 6756–6769 (2007).
Dietz, W. H. Jr Childhood obesity. Ann. N. Y. Acad. Sci. 499, 47–54 (1987).
West, A. C. & Johnstone, R. W. New and emerging HDAC inhibitors for cancer treatment. J. Clin. Invest. 124, 30–39 (2014).
Xiao, X. S. et al. High expression of p300 in human breast cancer correlates with tumor recurrence and predicts adverse prognosis. Chin. J. Cancer Res. 23, 201–207 (2011).
Fermento, M. E. et al. Inhibition of p300 suppresses growth of breast cancer. Role of p300 subcellular localization. Exp. Mol. Pathol. 97, 411–424 (2014).
Hou, X. et al. P300 promotes proliferation, migration, and invasion via inducing epithelial-mesenchymal transition in non-small cell lung cancer cells. BMC Cancer 18, 641 (2018).
Cai, L. Y. et al. Targeting p300/CBP atenuates hepatocellular carcinoma progression through epigenetic regulation of metabolism. Cancer Res. 81, 860–872 (2021).
Welti, J. et al. Targeting the p300/CBP axis in lethal prostate cancer. Cancer Discov. 11, 1118–1137 (2021).
van Gils, N. et al. The novel oral BET-CBP/p300 dual inhibitor NEO2734 is highly effective in eradicating acute myeloid leukemia blasts and stem/progenitor cells. Hemasphere 5, e610 (2021).
He, Z. X. et al. Current development of CBP/p300 inhibitors in the last decade. Eur. J. Med. Chem. 209, 112861 (2021).
Xiong, Y., Zhang, M. & Li, Y. Recent advances in the development of CBP/p300 bromodomain inhibitors. Curr. Med. Chem. 27, 5583–5598 (2020).
Giles F, Witcher M, Brown B. 429PNEO2734: a novel potent oral dual BET and P300/CBP inhibitor. Ann. Oncol. 29, viii140–viii141 (2018).
Bowers, E. M. et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem. Biol. 17, 471–482 (2010).
Oike, T. et al. C646, a selective small molecule inhibitor of histone acetyltransferase p300, radiosensitizes lung cancer cells by enhancing mitotic catastrophe. Radiother. Oncol. 111, 222–227 (2014).
Ono, H. et al. C646 inhibits G2/M cell cycle-related proteins and potentiates anti-tumor effects in pancreatic cancer. Sci. Rep. 11, 10078 (2021).
Dou, C. et al. P300 acetyltransferase mediates stiffness-induced activation of hepatic stellate cells into tumor-promoting myofibroblasts. Gastroenterology 154, 2209–2221 (2018).
Wang, Y. M. et al. Histone acetyltransferase p300/CBP inhibitor C646 blocks the survival and invasion pathways of gastric cancer cell lines. Int. J. Oncol. 51, 1860–1868 (2017).
He, H. et al. Selective p300 inhibitor C646 inhibited HPV E6-E7 genes, altered glucose metabolism and induced apoptosis in cervical cancer cells. Eur. J. Pharmacol. 812, 206–215 (2017).
Zheng, S. et al. Inhibiting p53 acetylation reduces cancer chemotoxicity. Cancer Res. 77, 4342–4354 (2017).
Gu, M. L. et al. An inhibitor of the acetyltransferases CBP/p300 exerts antineoplastic effects on gastrointestinal stromal tumor cells. Oncol. Rep. 36, 2763–2770 (2016).
Lasko, L. M. et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 550, 128–132 (2017).
Zhang, B., Chen, D., Liu, B., Dekker, F. J. & Quax, W. J. A novel histone acetyltransferase inhibitor A485 improves sensitivity of non-small-cell lung carcinoma cells to TRAIL. Biochem. Pharmacol. 175, 113914 (2020).
Zhou, X. R. et al. P300/CBP inhibition sensitizes mantle cell lymphoma to PI3Kdelta inhibitor idelalisib. Acta Pharmacol. Sin. 43, 457–469 (2022).
Lasko, L. M. et al. Inhibition of p300/CBP suppresses castration-resistant cancer. Cancer Discov. 7, 1215–1215 (2017).
Liu, J. et al. P300/CBP inhibition enhances the efficacy of programmed death-ligand 1 blockade treatment in prostate cancer. Oncogene 39, 3939–3951 (2020).
Durbin, A. D. et al. Ep300 selectively controls the enhancer landscape of MYCN-amplified neuroblastoma. Cancer Discov. 12, 730–751 (2022).
Peng, C. et al. A critical role for ZDHHC2 in metastasis and recurrence in human hepatocellular carcinoma. Biomed. Res. Int. 2014, 832712 (2014).
Sharma, C. et al. Protein acyltransferase DHHC3 regulates breast tumor growth, oxidative stress, and senescence. Cancer Res. 77, 6880–6890 (2017).
Qiu, N. et al. Artemisinin inhibits NRas palmitoylation by targeting the protein acyltransferase ZDHHC6. Cell Chem. Biol. 29, 530–537 (2022).
Sudo, H. et al. ZDHHC8 knockdown enhances radiosensitivity and suppresses tumor growth in a mesothelioma mouse model. Cancer Sci. 103, 203–209 (2012).
Oo, H. Z. et al. Overexpression of ZDHHC14 promotes migration and invasion of scirrhous type gastric cancer. Oncol. Rep. 32, 403–410 (2014).
Chen, S. et al. Targeting MC1R depalmitoylation to prevent melanomagenesis in redheads. Nat. Commun. 10, 877 (2019).
Yu, L. et al. Activation of a novel palmitoyltransferase ZDHHC14 in acute biphenotypic leukemia and subsets of acute myeloid leukemia. Leukemia 25, 367–371 (2011).
Bielawska, A. et al. Novel analogs of D-e-MAPP and B13. Part 2: signature effects on bioactive sphingolipids. Bioorg. Med. Chem. 16, 1032–1045 (2008).
Sangha, R. S. et al. An open-label, first-in-human, phase I trial of the safety and efficacy of daily PCLX-001. J. Clin. Oncol. 38, TPS3656 (2020).
Kallemeijn, W. W. et al. Validation and invalidation of chemical probes for the human N-myristoyltransferases. Cell Chem. Biol. 26, 892–900 (2019).
Elsey, J. et al. Palladium based nanoparticles for the treatment of advanced melanoma. Sci. Rep. 9, 3255 (2019).
Sangha, R. et al. Novel, first-in-Human, oral PCLX-001 treatment in a patient with relapsed diffuse large B-cell lymphoma. Curr. Oncol. 29, 1939–1946 (2022).
Beauchamp, E. et al. Targeting N-myristoylation for therapy of B-cell lymphomas. Nat. Commun. 11, 5348 (2020).
Ko, P. J. & Dixon, S. J. Protein palmitoylation and cancer. EMBO Rep. 19, e46666 (2018).
Lin, D. T. S., Davis, N. G. & Conibear, E. Targeting the Ras palmitoylation/depalmitoylation cycle in cancer. Biochem. Soc. Trans. 45, 913–921 (2017).
Chong, L. W., Tsai, C. L., Yang, K. C., Liao, C. C. & Hsu, Y. C. Targeting protein palmitoylation decreases palmitateinduced sphere formation of human liver cancer cells. Mol. Med. Rep. 22, 939–947 (2020).
Zhang, L. et al. HDAC1 knockdown inhibits invasion and induces apoptosis in non-small cell lung cancer cells. Biol. Chem. 399, 603–610 (2018).
Liu, X. et al. HDAC1 silencing in ovarian cancer enhances the chemotherapy response. Cell. Physiol. Biochem. 48, 1505–1518 (2018).
Chen, C. et al. The histone deacetylase HDAC1 activates HIF1alpha/VEGFA signal pathway in colorectal cancer. Gene 754, 144851 (2020).
Stojanovic, N. et al. HDAC1 and HDAC2 integrate the expression of p53 mutants in pancreatic cancer. Oncogene 36, 1804–1815 (2017).
Kim, J. K. et al. Targeted inactivation of HDAC2 restores p16INK4a activity and exerts antitumor effects on human gastric cancer. Mol. Cancer Res. 11, 62–73 (2013).
Yuan, J. H. et al. The histone deacetylase 4/SP1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology 54, 2025–2035 (2011).
Wang, X. C. et al. miR-433 inhibits oral squamous cell carcinoma (OSCC) cell growth and metastasis by targeting HDAC6. Oral. Oncol. 51, 674–682 (2015).
Bora-Singhal, N. et al. Novel HDAC11 inhibitors suppress lung adenocarcinoma stem cell self-renewal and overcome drug resistance by suppressing Sox2. Sci. Rep. 10, 4722 (2020).
Bi, L. et al. HDAC11 regulates glycolysis through the LKB1/AMPK signaling pathway to maintain hepatocellular carcinoma stemness. Cancer Res. 81, 2015–2028 (2021).
McClure, J. J., Li, X. & Chou, C. J. Advances and challenges of HDAC inhibitors in cancer therapeutics. Adv. Cancer Res. 138, 183–211 (2018).
Karagiannis, D. & Rampias, T. HDAC Inhibitors: dissecting mechanisms of action to counter tumor heterogeneity. Cancers 13, 3575 (2021).
Choy, E. et al. Phase 1 study of oral abexinostat, a histone deacetylase inhibitor, in combination with doxorubicin in patients with metastatic sarcoma. Cancer 121, 1223–1230 (2015).
Buggy, J. J. et al. CRA-024781: a novel synthetic inhibitor of histone deacetylase enzymes with antitumor activity in vitro and in vivo. Mol. Cancer Ther. 5, 1309–1317 (2006).
Bretz, A. C. et al. Domatinostat favors the immunotherapy response by modulating the tumor immune microenvironment (TIME). J. Immunother. Cancer 7, 294 (2019).
Putzer, S. et al. Reinstated p53 response and high anti-T-cell leukemia activity by the novel alkylating deacetylase inhibitor tinostamustine. Leukemia 34, 2513–2518 (2020).
Tsimberidou, A. M. et al. Preclinical development and first-in-human study of KA2507, a selective and potent inhibitor of histone deacetylase 6, for patients with refractory solid tumors. Clin. Cancer Res. 27, 3584–3594 (2021).
Booth, S. W. et al. A phase 2a cohort expansion study to assess the safety, tolerability, and preliminary efficacy of CXD101 in patients with advanced solid-organ cancer expressing HR23B or lymphoma. BMC Cancer 21, 851 (2021).
Olsson, A. et al. Tasquinimod triggers an early change in the polarization of tumor associated macrophages in the tumor microenvironment. J. Immunother. Cancer 3, 53 (2015).
Vujic, I. et al. Acyl protein thioesterase 1 and 2 (APT-1, APT-2) inhibitors palmostatin B, ML348 and ML349 have different effects on NRAS mutant melanoma cells. Oncotarget 7, 7297–7306 (2016).
Xu, J. et al. Inhibiting the palmitoylation/depalmitoylation cycle selectively reduces the growth of hematopoietic cells expressing oncogenic Nras. Blood 119, 1032–1035 (2012).
Adibekian, A. et al. Confirming target engagement for reversible inhibitors in vivo by kinetically tuned activity-based probes. J. Am. Chem. Soc. 134, 10345–10348 (2012).
Potts, M. B. et al. Mode of action and pharmacogenomic biomarkers for exceptional responders to didemnin B. Nat. Chem. Biol. 11, 401–408 (2015).
Rebecca, V. W. et al. A unified approach to targeting the lysosome’s degradative and growth signaling roles. Cancer Discov. 7, 1266–1283 (2017).
Remsberg, J. R. et al. ABHD17 regulation of plasma membrane palmitoylation and N-Ras-dependent cancer growth. Nat. Chem. Biol. 17, 856–864 (2021).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Delmore, J. E. et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011).
Mirguet, O. et al. From ApoA1 upregulation to BET family bromodomain inhibition: discovery of I-BET151. Bioorg. Med. Chem. Lett. 22, 2963–2967 (2012).
Lai, J., Liu, Z., Zhao, Y., Ma, C. & Huang, H. Anticancer effects of I-BET151, an inhibitor of bromodomain and extra-terminal domain proteins. Front. Oncol. 11, 716830 (2021).
Fiorentino, F. P. et al. BET-inhibitor I-BET762 and PARP-inhibitor talazoparib synergy in small cell lung cancer cells. Int. J. Mol. Sci. 21, 9595 (2020).
Ozer, H. G. et al. BRD4 profiling identifies critical chronic lymphocytic leukemia oncogenic circuits and reveals sensitivity to PLX51107, a novel structurally distinct BET inhibitor. Cancer Discov. 8, 458–477 (2018).
Brandts, J. & Ray, K. K. Apabetalone - BET protein inhibition in cardiovascular disease and Type 2 diabetes. Future Cardiol. 16, 385–395 (2020).
Ray, K. K. et al. Effect of apabetalone added to standard therapy on major adverse cardiovascular events in patients with recent acute coronary syndrome and type 2 diabetes: a randomized clinical trial. J. Am. Med. Assoc. 323, 1565–1573 (2020).
Maneiro, M. A. et al. Antibody-protac conjugates enable HER2-dependent targeted protein degradation of BRD4. ACS Chem. Biol. 15, 1306–1312 (2020).
Raina, K. et al. Protac-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc. Natl Acad. Sci. USA 113, 7124–7129 (2016).
Zhang, X. et al. Ultradeep palmitoylomics enabled by dithiodipyridine-functionalized magnetic nanoparticles. Anal. Chem. 90, 6161–6168 (2018).
Deng, W. et al. GPS-PAIL: prediction of lysine acetyltransferase-specific modification sites from protein sequences. Sci. Rep. 6, 39787 (2016).
Ning, W. et al. HybridSucc: a hybrid-learning architecture for general and species-specific succinylation site prediction. Genomics Proteom. Bioinforma. 18, 194–207 (2020).
Ren, J. et al. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 21, 639–644 (2008).
Liu, B. et al. Identification and characterization of propionylation at histone H3 lysine 23 in mammalian cells. J. Biol. Chem. 284, 32288–32295 (2009).
Chai, X. et al. Quantitative acetylome analysis reveals histone modifications that may predict prognosis in hepatitis B-related hepatocellular carcinoma. Clin. Transl. Med. 11, e313 (2021).
Wang, B. et al. SIRT2-dependent IDH1 deacetylation inhibits colorectal cancer and liver metastases. EMBO Rep. 21, e48183 (2020).
Acknowledgements
This work was supported by grants from the National Key R&D Program of China (2022YFC2504003, 2017YFA0205400), National Natural Science Foundation of China (82273947, 81973344 to F.H. and 81903636 to S.S.), CAMS Innovation Fund for Medical Sciences (CIFMS, 2021-I2M-1-021 to F.H. and S.S.; 2022-I2M-JB-011 to F.H.), Beijing Outstanding Young Scientist Program (BJJWZYJH01201910023028) and the CAMS Central Public-Interest Scientific Institution Basal Research Fund (2018PT35004). We thank Figdraw (www.figdraw.com) for the assistance in creating Figs. 4–6.
Author information
Authors and Affiliations
Contributions
F.H. designed the project. S.S. and F.H organized, wrote, and revised the manuscript. S.S. and J.L. summarized the literature. All authors have read and approved the final article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shang, S., Liu, J. & Hua, F. Protein acylation: mechanisms, biological functions and therapeutic targets. Sig Transduct Target Ther 7, 396 (2022). https://doi.org/10.1038/s41392-022-01245-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41392-022-01245-y
This article is cited by
-
Mechanisms and functions of protein S-acylation
Nature Reviews Molecular Cell Biology (2024)
-
Transcriptional Expression of Histone Acetyltransferases and Deacetylases During the Recovery of Acute Exercise in Mouse Hippocampus
Journal of Molecular Neuroscience (2024)
-
A glimpse into novel acylations and their emerging role in regulating cancer metastasis
Cellular and Molecular Life Sciences (2024)
-
Unveiling the role of GAS41 in cancer progression
Cancer Cell International (2023)
-
Lipid metabolic reprogramming in tumor microenvironment: from mechanisms to therapeutics
Journal of Hematology & Oncology (2023)