Abstract
Multiple myeloma (MM) is the second most common hematological malignancy. N6-methyladenosine (m6A) is the most abundant RNA modification. YTH domain-containing family protein 2 (YTHDF2) recognizes m6A-cotaining RNAs and accelerates degradation to regulate cancer progression. However, the role of YTHDF2 in MM remains unclear. We investigated the expression levels and prognostic role of YTHDF2 in MM, and studied the effect of YTHDF2 on MM proliferation and cell cycle. The results showed that YTHDF2 was highly expressed in MM and was an independent prognostic factor for MM survival. Silencing YTHDF2 suppressed cell proliferation and caused the G1/S phase cell cycle arrest. RNA immunoprecipitation (RIP) and m6A-RIP (MeRIP) revealed that YTHDF2 accelerated EGR1 mRNA degradation in an m6A-dependent manner. Moreover, overexpression of YTHDF2 promoted MM growth via the m6A-dependent degradation of EGR1 both in vitro and in vivo. Furthermore, EGR1 suppressed cell proliferation and retarded cell cycle by activating p21cip1/waf1 transcription and inhibiting CDK2-cyclinE1. EGR1 knockdown could reverse the inhibited proliferation and cell cycle arrest upon YTHDF2 knockdown. In conclusion, the high expression of YTHDF2 promoted MM cell proliferation via EGR1/p21cip1/waf1/CDK2-cyclin E1 axis-mediated cell cycle transition, highlighting the potential of YTHDF2 as an effective prognostic biomarker and a promising therapeutic target for MM.

Similar content being viewed by others
Introduction
Multiple myeloma (MM) is the second most common hematological malignancy characterized by the clonal proliferation of malignant plasm cells in the bone marrow [1]. Initiation of MM is characterized by monoclonal gammopathy of unknown significance (MGUS) and smoldering MM (SMM), finally developing into the extramedullary MM (EMM) and plasma cell leukemia (PCL). Treatment of MM involves the use of proteasome inhibitor-based regimen, and is supplemented with immunotherapy, including monoclonal antibodies and chimeric antigen receptor-engineered T cells [2]. However, MM remains incurable with high mortality rate. Therefore, it is urgent to understand the pathogenesis of MM and explore novel therapeutic targets.
N6-methyladenosine (m6A) is the most abundant RNA modification among more than 160 types of chemical post-transcriptional modifications [3,4,5]. M6A sites mainly occur at the consensus motif of RRACH (R = G/A, H = A/C/U) in 3’untranslated regions (3’UTR), around stop codons, within internal long exons and precursor mRNAs [6, 7]. Notably, the process of m6A RNA methylation is reversible: m6A is installed by “writers”, removed by “erasers”, and recognized by “readers”. Methyltransferase-like 3 (METTL3) and METTL14, key components of methyltransferase complex, cooperating with adapter proteins, such as WT1-associated protein (WTAP), conduct the methyl transfer. On the other hand, fat mass and obesity-associated protein (FTO) and α-ketoglutarate-dependent dioxygenase homolog 5 (ALKBH5) are RNA demethylases responsible for the removal of m6A modification [8]. Additionally, readers can recognize specific m6A sites and regulate RNA splicing, miRNA processing, RNA export, degradation and translation [9, 10].
YTH domain-containing family protein 2 (YTHDF2) is the first identified m6A reader. YTHDF2 can recognize m6A-cotaining RNA and accelerate the degradation process to regulate many biological processes, such as viral infection, stem cell development and cancer progression [11,12,13]. Paris et al. [14] demonstrated that YTHDF2 played an essential role in the initiation and development of leukemia stem cells. Moreover, YTHDF2 was shown to promote epithelial-mesenchymal transition and AKT phosphorylation to support prostate cancer progression [15]. However, the role of YTHDF2 in MM progression remains largely unclear. In this study, we observed the increased expression of YTHDF2 in MM, and showed that YTHDF2 was an independent prognostic factor for overall survival (OS) in MM. Mechanistically, this study revealed that YTHDF2 promoted MM cell proliferation through the EGR1/p21cip1/waf1/CDK2-Cyclin E1 axis-mediated cell cycle transition. These findings suggested that YTHDF2 could be a potential biomarker and promising therapeutic target in MM.
Methods and Materials
Cell lines and cell culture
Human myeloma cell lines MM.1S, RPMI-8226, NCI-H929 were purchased from the American Type Culture Collection (USA), OPM2 and U266 were obtained from Professor Jinsong Hu of Xi’an Jiaotong University Health Science Center (Xian, Shaanxi, China). MM cell lines were maintained in RPMI-1640 (Hyclone, Logan, UT, USA) with 10% fetal bovine serum (FBS) (Biological Industries, Kibbutz Beit Haemek, Israel), penicillin (10,000 U/L, BioSharp, Hefei, China) and streptomycin (100 mg/L, BioSharp, Hefei, China). All cells were incubated at 37 °C in a humidified atmosphere with 5% CO2.
Patients
Bone marrow specimens were obtained from newly diagnosed MM at the Department of Hematology, the Second Affiliated Hospital of Xi’an Jiaotong University, from 2018 to 2022. Peripheral blood-derived mononuclear cells from healthy doners were used as control. The clinical samples used in this study to identify the expression of YTHDF2 were supervised and granted by the Ethics Committee of the Second Affiliated Hospital of Xi’an Jiaotong University (2015186).
RNA immunoprecipitation (RIP) assay
RIP assay was performed using the BersinBioTM RNA Immunoprecipitation Kit (BersinBio, Guangzhou, China) according to the manufacturers’ instruction. Polysome lysis buffer was used to lyse cells, and the cell lysate was divided into anti-YTHDF2, anti-IgG (1 mg/ml, Cell Signaling Technology) and input samples. The cell lysates were incubated with pre-conjugated protein A/G magnetic beads with 5 μg of specific antibodies overnight at 4 °C. After washing, the lysates were digested with Proteinase K for 1 h at 55 °C, and the RNA bound to immunoprecipitated proteins was purified by phenol: chloroform: isoamyl alcohol (25:24:1). Realtime-qPCR was performed to measure the target RNA expression levels. All primer sequences are shown in Supplementary Table 1.
m6A-RNA immunoprecipitation (MeRIP) assay
MeRIP assay was performed using the BersinBioTM MeRIP Kit (BersinBio, Guangzhou, China) according to the manufacturers’ instruction. Total RNA was extracted using TRIzol reagent. Then, the RNA was sheared into about 300-nucleotide fragments. Next, RNA was incubated with 5 μg anti-m6A antibody (68055-1-Ig, Proteintech) or IgG for 2 h at 4 °C. Protein A/G magnetic beads were mixed with the antibody-treated RNA in IP buffer for 2 h at 4 °C. Then, the bound RNAs were washed and eluted with Proteinase K and elution buffer for 1 h at 55 °C. Finally, RNA bound to immunoprecipitated proteins was purified by phenol: chloroform: isoamyl alcohol (25:24:1). Realtime-qPCR was performed to measure the methylated RNA expression levels.
RNA stability assay
Cells were treated with Actinomycin D (5 μg/ml, MCE, USA) for 0, 10, 20, 40, 60 min or longer. Total RNA was then extracted by TRIzol. The target RNA was measured by Realtime-qPCR. 18 s rRNA was used as the endogenous standard control for mRNA normalization. The mRNA degradation rate Kdacay and RNA lifetime (t1/2) can be calculated using the equations [16]: Nt/N0 = e – Kdecay*t, t1/2 = ln2/Kdecay.
Animal experiment
Animal experiment was supervised and granted by the Ethic Committee of Xi’an Jiaotong University Health Science Center (2022-1497). Male BALB/c nude mice (4 weeks old) were use in this study. The mice were fed in specific pathogen-free facilities. RPMI-8226 cells transfected with negative control (NC) or shYTHDF2 or LV-oeYTHDF2 were injected into the left and right flank of mice. Tumor volume was recorded every 3 days, and calculated by the formula: (width2 × length × 0.5). Mice were sacrificed 4 weeks after injection or when the tumor diameter was more than 15 mm. Tumors were sent for further immunohistochemistry (IHC) staining as previously described [17].
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed by the ChIP assay Kit (Beyotime, China) according to the manufacturer’s instruction. Briefly, one million MM cells were cross-linked with 1% formaldehyde for 10 min at 37 °C. Subsequently, 0.125 M glycine was added to terminate the reaction, and cells were lysed on ice by lysis buffer. Chromatin DNA was sonicated to obtained 200-1000 bp fragments, which was followed by incubation with anti-EGR1 antibody or IgG at 4 °C overnight. EGR1-bound DNA fragments were precipitated by Protein A + G Agarose/Salmon sperm DNA. After de-crosslink and purification, qPCR and agarose gel electrophoresis were employed to analyze the precipitated DNA. The EGR1-binding sequence of p21cip1/waf1 promoter was predicted by JASPAR database (https://jaspar.genereg.net/) [18].
Statistical analysis
Three dependent biological replicates were used in each experiment. Mann-Whitney test and Student’s t test were used to compare the difference in two groups as appropriate. One-way ANOVA followed by Bonferroni post hoc comparison were employed to compare the difference of more than two subgroups. P < 0.05 was considered statistically significant if not specified. All statistical tests were two-sided.
Results
YTHDF2 is highly expressed in MM patients
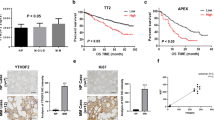
To explore the association between YTHDF2 expression and MM, we performed a comprehensive analysis of the development of MM. The results revealed significantly higher expression of YTHDF2 in MM than in both MGUS (P = 0.0008) and SMM (P = 0.0002) (Fig. 1A). Moreover, the expression of YTHDF2 was higher in PCL than in MM (P = 0.0210, Fig. 1B). Furthermore, higher Durie-Salmon system (DSS), International Staging System (ISS) and Revised-ISS (R-ISS) stages were associated with increased expression of YTHDF2 in MM patients (Fig. 1C–E). There was also a significant increase in the expression of YTHDF2 in relapsed MM patients (Supplementary Fig. 1A–C) and those with bortezomib (BTZ) resistance (Supplementary Fig. 1D, E). Overall, these findings showed that YTHDF2 had an oncogenic role in MM, and that was positively correlated with advanced-stage MM, recurrence and resistance to bortezomib.
A–E the YTHDF2 expression in MM progression: normal plasma cell (NPC), MGUS, SMM, MM (A); PCL (B); DSS (C), ISS (D) and R-ISS stages (E). F Kaplan-Meier survival analysis showing the OS of MM patients having the low YTHDF2 expression (blue) and high YTHDF2 expression (red). G Nomogram predicting 1-, 3-, 5-year survival of MM patients. (H) Calibration plots of the nomogram for 1-year, 2-year and 3-year OS. (I) Kaplan-Meier survival analysis for total points of Nomogram. J ROC curve for the nomogram prognosis system. K The expression of YTHDF2 in MM cell lines and normal plasm cells by RT-qPCR. L–N The expression of YTHDF2 in normal plasma cells and MM patients grouped by ISS and R-ISS stages. Data are mean ± SD values. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s. not significant.
The expression of YTHDF2 is an independent factor for MM survival
The results further showed that the OS, event-free survival (EFS) and progression-free survival (PFS) of MM patients with higher levels of YTHDF2 expression was shorter than that of patients with low YTHDF2 expression (Fig. 1F, Supplementary Fig. 1F–J). In addition, univariate and multivariate Cox regression analyses revealed that the expression level of YTHDF2 was an independent prognostic factor for MM survival (P = 0.02, Table 1). We further combined YTHDF2 expression with other independent prognostic factors and constructed a nomogram to calculate the total points (Fig. 1G). The calibration curves illustrated great agreement between the predicted and actual 1-, 2- and 3- year survival probabilities (Fig. 1H). Notably, MM patients with higher total points showed significantly poorer OS than those with lower total points (P < 0.0001, Fig. 1I). The area under ROC curve (AUC) of the total points for 1-, 2- and 3- year survival probabilities was 0.7858 (95%CI: 0.6934 to 0.8773, P < 0.0001), 0.7763 (95%CI: 0.7013 to 0.8513, P < 0.0001), and 0.7629 (95%CI: 0.6982 to 0.8275, P < 0.0001), respectively (Fig. 1J). ROC analysis and decision curve analysis (DCA) further showed that the inclusion of YTHDF2 could improve the evaluation accuracy of MM prognosis (Supplementary Fig. 1K–N).
Next, we examined YTHDF2 expression in both MM cell lines and patients. The findings revealed that YTHDF2 was highly expressed in MM cell lines and patients compared to normal plasm cells (Fig. 1K, L). In addition, there was increased expression of YTHDF2 in MM patients with more advanced ISS and R-ISS stage (Fig. 1M, N). Collectively, the results suggested that YTHDF2 could be a potential biomarker for predicting MM prognosis, and the combination of YTHDF2 with clinical features could effectively improve the accuracy of prognosis evaluation.
Silencing YTHDF2 inhibits MM cell proliferation, and causes G1/S phase cell cycle arrest
To further explore the role of YTHDF2 in MM, we downregulated YTHDF2 expression in MM cell lines RPMI-8226 and NCI-H929 using specific siRNAs (Fig. 2A, B). The results showed the knockdown of YTHDF2 had no significant effect on the expression levels of the other m6A regulators, METTL3, FTO, ALKBH5, YTHDF1, and YTHDF3 (P > 0.05, Supplementary Fig. 2A–D). We observed that the knockdown of YTHDF2 significantly inhibited cell viability (Fig. 2C, D) and cell proliferation (Fig. 2E, F). Cell cycle analysis further revealed that the downregulation of YTHDF2 in RPMI-8226 and NCI-H929 resulted in the increase of G1 phase cells and reduction of S phase cells, while no significant difference was observed in the G2 phase, suggesting possible G1/S phase arrest (Fig. 2G, H).
A, B Validation of YTHDF2 knockdown at both mRNA and protein levels in RPMI-8226 and NCI-H929 MM cell lines. C, D Cell viability was detected by CCK8 assays in RPMI-8226 and NCI-H929 cells transfected with siNC and siYTHDF2. E, F Cell proliferation was measured by EdU assay in RPMI-8226 and NCI-H929 cells transfected with siRNAs. G, H Cell cycle was detected by PI-staining in RPMI-8226 and NCI-H929 cells transfected with siNC and siYTHDF2. Data are mean ± SD values. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s. not significant.
To reinforce our findings, MM cell lines were transfected with lentivirus vectors carrying YTHDF2-shRNA and scrambled shRNA. The findings similarly showed that downregulation of YTHDF2 significantly suppressed cell proliferation and caused cell cycle arrest (Supplementary Fig. 2E–H).
YTHDF2 enhances the degradation of EGR1 mRNA in an m6A-dependent manner
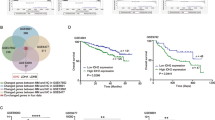
Given that YTHDF2 could promote the degradation of m6a-containing transcripts, we analyzed the significantly downregulated genes in MM patients relative to healthy individuals based on the GSE47552 dataset. We combined these downregulated genes with PAR-CLIP-seq results of YTHDF2 from POSTAR3 online database (http://lulab.life.tsinghua.edu.cn/postar/) [19], finally obtaining 388 genes (Fig. 3A, Supplementary Table 2). Next, Gene Ontology and pathway enrichment analyses were conducted using the online Metascape tool (https://metascape.org/gp) [20], which showed that YTHDF2 was most involved in the cell cycle (Fig. 3B, Supplementary Table 3). Among these genes related to cell cycle, early growth response factor 1 (EGR1) was the most downregulated one (fold change = −4.14, Supplementary Table 2), hence selected for further validation. Furthermore, we found a significant negative correlation between the expression of YTHDF2 and EGR1 (Supplementary Fig. 3A–C). Compared with normal plasm cells, the expression of EGR1 was lower in MGUS, SMM and MM (Fig. 3C). In addition, there was a significant decrease in the expression of EGR1 in PCL and MM patients with a higher ISS stage as well as in relapsed MM patients (Fig. 3D–F). It was also shown that MM patients with low expression of EGR1 had remarkably poorer OS (Fig. 3G). Moreover, we confirmed the downregulation of EGR1 in MM cell lines compared with normal plasm cells (Fig. 3H).
A The Venn plot showed 388 overlapped genes between significantly downregulated genes in MM from GSE47552 and potential RNA targets of YTHDF2 from POSTAR database. B The enrichment analysis was conducted by Metascape. C–F The expression of EGR1 in the development of MM: NPC, MGUS, SMM, MM (C); PCL (D); ISS stage (E) and relapsed MM patients (F). G Kaplan-Meier survival analysis showing the OS for MM patients having low EGR1 expression (blue) and high EGR1 expression (red) in the GSE24080 dataset. H The expression of EGR1 in MM cell lines and normal plasm cells. I, J The mRNA and protein levels of EGR1 upon YTHDF2 knockdown in MM cell lines RPMI-8226 and NCI-H929. K RNA immunoprecipitation assays of EGR1 transcripts in YTHDF2-bound mRNAs in RPMI-8226 and NCI-H929 cells. L RNA stability analysis of EGR1 after actinomycin D treatment in RPMI-8226 cells transfected with siNC and siYTHDF2. M The putative m6A sites of EGR1 mRNA were predicted by POSTAR3. N Methylated RNA immunoprecipitation of EGR1 transcripts in RPMI-8226 and NCI-H929 transfected with siNC and siYTHDF2. Data are mean ± SD values. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s. not significant.
Further, the ability of YTHDF2 to regulate the expression of EGR1 in an m6A-dependent manner was investigated. The findings showed that knockdown of YTHDF2 significantly upregulated the expression of EGR1 (Fig. 3I, J). In addition, RIP-qPCR verified the interaction between the YTHDF2 protein and EGR1 mRNA (Fig. 3K). The half-life of EGR1 mRNA was also significantly prolonged in YTHDF2-downregulated MM cells (28.95 min versus 35.44 min, P = 0.0377), suggesting that YTHDF2 could decrease the stability of EGR1 mRNA (Fig. 3L). Then, several potential m6A sites for EGR1 mRNA were discovered using SRAMP [21] (http://www.cuilab.cn/sramp/, Fig. 3M), and MeRIP-qPCR was performed. Knockdown of YTHDF2 significantly increased m6A modification of EGR1 mRNA in MM cells (Fig. 3N). Taken together, these results suggested that EGR1 mRNA was a direct target of YTHDF2, and YTHDF2 could accelerate EGR1 mRNA degradation in an m6A-dependent manner in MM.
EGR1 inhibits cell proliferation and causes cell cycle arrest by transcriptionally regulating p21cip1/waf1
To further validate the role of EGR1 in MM, EGR1 was silenced in MM cell lines (Fig. 4A, B). The EdU assay showed that silencing EGR1 significantly enhanced cell proliferation (Fig. 4C, D). Moreover, cell cycle analysis revealed that the downregulation of EGR1 caused an increase in S phase cells and a decrease in G1 phase cells, suggesting the promotion of the G1/S phase (Fig. 4E, F).
A, B The mRNA and protein levels of EGR1 and p21cip1/waf1 upon EGR1 knockdown in MM cell lines RPMI-8226 and NCI-H929. C, D Cell proliferation was measured by EdU assay in RPMI-8226 and NCI-H929 cells transfected with siNC or siEGR1. (E, F) Cell cycle was detected by PI staining in RPMI-8226 and NCI-H929 cells transfected with siNC and siEGR1. G The EGR1-binding motif of p21 promoter (above) was predicted by JASPAR database. H, I ChIP assay was analyzed by RT-qPCR and agarose gel electrophoresis. J Luciferase reporter assays of wt- and mut-p21cip1/waf1 promoter sequences following EGR1 knockdown or YTHDF2 overexpression. Data are mean ± SD values. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s. not significant.
The cyclin-dependent kinase inhibitor 1 A (CDKN1A), also named p21cip1/waf1, is a well-studied tumor suppressor, known for mediating cell cycle arrest [22]. Previous studies have shown that EGR1, as a transcriptional factor, can promote the activation of p21cip1/waf1 in glioma [23, 24], breast cancer [25], melanoma [26] and gastric cancer [27]. In addition, a positive correlation between EGR1 and p21cip1/waf1 expression was found in MM (Supplementary Fig. 3D, E). Therefore, we hypothesized that EGR1 could suppress the proliferation of MM cells and cause cell cycle arrest by activating p21cip1/waf1 transcription. Expectedly, the knockdown of EGR1 significantly decreased the expression of p21cip1/waf1 (Fig. 4A, B). To further assess whether EGR1 was involved in the transcriptional activation of p21cip1/waf1 in MM cells, we predicted the potential EGR1-binding motif on the p21cip1/waf1 promoter using JASPAR (Fig. 4G). The ChIP-qPCR assay confirmed the interaction between EGR1 and p21cip1/waf1 promoter (Fig. 4H, I). In addition, we constructed luciferase reporter plasmids with wild type (wt) or mutant (mut) sequences, and found that luciferase activity in the wt-luciferase reporter was significantly decreased upon both the knockdown of EGR1 and the overexpression of YTHDF2. However, no significant difference was observed in the mut-luciferase reporter (Fig. 4J). Collectively, these findings suggested that EGR1 could suppress the proliferation of MM cells and retard cell cycle by transcriptionally activating p21cip1/waf1.
Silencing YTHDF2 suppresses cell proliferation and cell cycle via the EGR1/ p21cip1/waf1/CDK2-Cyclin E1 axis
Previous research demonstrated that cyclin-dependent kinase 2 (CDK2)-Cyclin E1 was involved in p21cip1/waf1-mediated G1/S cell cycle transition [22]. Therefore, we speculated that YTHDF2 could regulate cell proliferation and cell cycle via the EGR1/p21cip1/waf1/CDK2-Cyclin E1 axis in MM. As expected, there was an increase in p21cip1/waf1 expression, while CDK2 and Cyclin E1 expression decreased following YTHDF2 knockdown in MM cell lines (Fig. 5A). Conversely, knockdown of EGR1 caused the downregulation of p21cip1/waf1, but resulted in the upregulation of CDK2 and cyclin E1 (Fig. 5B).
A, B The protein levels of the YTHDF2/EGR1/ p21cip1/waf1/CDK2-Cyclin E1 axis upon YTHDF2 knockdown (A) and EGR1 knockdown (B) in MM cell lines RPMI-8226 and NCI-H929. C, D Realtime-qPCR results of the EGR1 (C) and p21cip1/waf1 (D) mRNA expression upon silencing both YTHDF2 and EGR1 in RPMI-8226 and NCI-H929. E The protein levels of the YTHDF2/EGR1/ p21cip1/waf1/CDK2-Cyclin E1 axis upon silencing both YTHDF2 and EGR1 in RPMI-8226 and NCI-H929. F, G Cell cycle was detected by PI staining in RPMI-8226 and NCI-H929 cells simultaneously transfected with siYTHDF2 and siEGR1. H, I Cell proliferation was measured by EdU assay in RPMI-8226 and NCI-H929 cells simultaneously transfected with siYTHDF2 and siEGR1. Data are mean ± SD values. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s. not significant.
To further investigate the YTHDF2/EGR1/p21cip1/waf1/CDK2-Cyclin E1 axis in MM, we simultaneously silenced YTHDF2 and EGR1 in MM cell lines. Knockdown of EGR1 not only reversed the increased expression of EGR1 and p21cip1/waf1, but also the decreased expression of CDK2 and Cyclin E1 caused by YTHDF2 silencing (Fig. 5C–E). Additionally, cell cycle analysis showed that EGR1 knockdown significantly reversed the G1/S phase arrest induced by YTHDF2 knockdown (Fig. 5F, G). The EdU assays also showed that silencing EGR1 significantly enhanced cell proliferation in YTHDF2-knockdown MM cells (Fig. 5H, I). Overall, these results demonstrated that YTHDF2 could promote cell proliferation and G1/S transition via the EGR1/p21cip1/waf1/CDK2-Cyclin E1 axis in MM.
Knockdown of YTHDF2 suppresses tumor growth in MM xenograft models
To assess the effect of YTHDF2 on MM proliferation in vivo, we subcutaneously injected RPMI-8226 cells transfected with shNC or shYTHDF2 into nude mice. Generally, tumors from shYTHDF2-transfected RPMI-8226 cells were visually smaller than those from shNC-transfected RPMI-8226 cells (Fig. 6A, B). Knockdown of YTHDF2 significantly decreased both tumor volume (P = 0.0304, Fig. 6C) and tumor weight (P = 0.0496, Fig. 6D). IHC results confirmed the decreased expression of YTHDF2, CDK2, Cyclin E1, and Ki-67, and the increased expression of EGR1 and p21cip1/waf1 in YTHDF2-downreuglated xenografts (Fig. 6E, F; Supplementary Fig. 2I). Taken together, these data confirmed the oncogenic role of YTHDF2 in promoting MM proliferation via the EGR1/p21cip1/waf1/CDK2-Cyclin E1 axis in vivo.
A RPMI-8226 cells transfected with shNC or shYTHDF2 were injected in the left or right flanks of BALB/c nude mice (n = 4). B Mice were sacrificed when tumor diameter was more than 15 mm, xenograft tumors were completely dissected. C Tumor volume was recorded every 3 days and calculated by the formula: (width2 × length ×0.5). (D) Tumor weight was recorded after dissection. E Representative hematoxylin-eosin (H&E) staining. F Representative IHC staining of YTHDF2, EGR1, p21cip1/waf1, CDK2, Cyclin E1 and Ki-67 in tumors from negative control and YTHDF2-downregulated nude mice. Data are mean ± SD values. *P < 0.05.
Overexpression of YTHDF2 promotes MM growth in vitro and in vivo
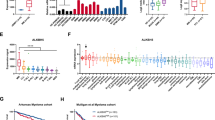
The findings further showed that the overexpression of YTHDF2 by lentiviral transduction (LV-oeYTHDF2) significantly reduced the expression of EGR1 and p21, but enhanced the expression of CDK2 and Cyclin E1 (Fig. 7A, B). In addition, the upregulation of YTHDF2 significantly promoted cell proliferation and the progression of cell cycle (Fig. 7C–F). The half-life of EGR1 mRNA was significantly shortened in MM cells overexpressing YTHDF2 (29.82 min versus 18.216 min; Fig. 7G). The MeRIP-qPCR also showed that overexpression of YTHDF2 significantly decreased the m6A modification of EGR1 mRNA (Fig. 7H).
A, B The expression of the YTHDF2/EGR1/ p21cip1/waf1/CDK2-Cyclin E1 in RPMI-8226 and NCI-H929 transfected with LV-NC and LV-oeYTHDF2. C, D Cell proliferation was measured by EdU assay in RPMI-8226 and NCI-H929 transfected with LV-NC and LV-oeYTHDF2. E, F Cell cycle was detected by PI-staining in RPMI-8226 and NCI-H929 transfected with LV-NC and LV-oeYTHDF2. G RNA stability analysis of EGR1 after actinomycin D treatment upon YTHDF2 overexpression. H Methylated RNA immunoprecipitation of EGR1 transcripts in YTHDF2-upregulated MM cells. I RPMI-8226 cells transfected with LV-NC or LV-oeYTHDF2 were injected in the left or right flanks of BALB/c nude mice (n = 6). Mice were sacrificed when tumor diameter was more than 15 mm, xenograft tumors were completely dissected. J Tumor volume was recorded every 3 days and calculated by the formula: (width2 × length ×0.5). (K) Tumor weight was recorded after dissection. L Representative H&E staining in tumors from LV-NC (above) and LV-oeYTHDF2 (below) mice. M Representative IHC staining of YTHDF2, EGR1, p21cip1/waf1, CDK2, Cyclin E1 and Ki-67 in tumors from LV-NC and LV-oeYTHDF2 nude mice. Data are mean ± SD values. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s. not significant.
Furthermore, we subcutaneously injected RPMI-8226 cells transfected with LV-NC and LV-oeYTHDF2 into nude mice, and observed that tumors from the LV-oeYTHDF2 group were bigger than those from LV-NC group (Fig. 7I). Additionally, the overexpression of YTHDF2 significantly increased both tumor volume (P = 0.0063, Fig. 7J) and tumor weight (P = 0.0076, Fig. 7K). IHC results confirmed the decreased expression of EGR1 and p21cip1/waf1, and increased expression of YTHDF2, CDK2, Cyclin E1, and Ki-67 in LV-oeYTHDF2 xenografts compared with the LV-NC group (Fig. 7L, M, Supplementary Fig. 2J).
Discussion
MM is a highly heterogeneous and genetically complicated hematological malignancy. According to existing evidence, MM initiation and progression is mainly dependent on a series of chromosomal and genetic alterations, such as IgH translocation, hyperdiploidy, amplification of the 1q chromosome, KRAS and TP53 mutations [28]. Accurate risk stratification and novel therapeutic regimens have significantly improved the survival of MM patients. However, the considerable clonal heterogeneity of MM makes it challenging to implement individualized and targeted therapy [29]. In the recent years, epigenetic abnormalities, such as RNA modifications along with DNA and histone methylation, have been demonstrated to contribute to MM heterogeneity [30]. Notably, m6A is the most common form of RNA modification, and recent studies have shown that m6A regulators are dysregulated in multiple cancers. Nevertheless, the role of m6A modification in MM oncogenesis remains unknown. In this study, the expression of YTHDF2 in MM patients was shown to be significantly higher than that in healthy individuals. Additionally, the knockdown of YTHDF2 significantly inhibited the proliferation of MM cells, caused cell cycle G1/S cycle arrest in vitro, and suppressed tumor growth in vivo. However, the overexpression of YTHDF2 reversed these effects. Mechanistically, YTHDF2 recognized and accelerated the degradation of EGR1 mRNA in an m6A-dependent manner. Moreover, without the transcriptional activation of EGR1, the reduced p21cip1/waf1 expression reversed its inhibitory effect on CDK2-Cyclin E1 and promoted G1/S phase transition to facilitate MM growth.
YTHDF2 is an important m6A reader in regulating RNA degradation. In the human YTHDF2-YTH domain, the aromatic cage composed of W486, W432, and W491 of the hydrophobic pocket was shown to recognize and trap m6A sites [31, 32]. Then, YTHDF2 could induce RNA decay through the deadenylation-dependent [33] and endonuclease-mediated pathways [34]. Since YTHDF2 functions to regulate gene expression post-transcriptionally, accumulating evidence suggests that YTHDF2 is involved in oncogenesis. For instance, Li et al. [35] revealed that YTHDF2 was highly expressed in lung adenocarcinoma and promoted tumor growth by degrading AXIN1 mRNA and activating the Wnt/beta-catenin signaling pathway. However, the biofunction of YTHDF2 is heterogeneous in cancers, with some studies demonstrating its tumor-inhibiting function. In hepatocellular carcinoma, Zhong et al. [36] revealed that HIF-1α could suppress YTHDF2 expression and activate the ERK/MEK signaling pathway to promote tumor growth by stabilizing EGFR mRNA. However, little research emphasizes the role of YTHDF2 in MM. Recently, Hua et al. [37] reported increased expression of YTHDF2 in MM, which was associated with poor prognosis. They demonstrated that YTHDF2 promoted MM proliferation, consistent with our results, further strengthening the carcinogenic role of YTHDF2 in MM. They also confirmed that YTHDF2 promoted MM progression by degrading STAT5 mRNA and activating the MP2K2/p-ERK signaling pathway. In the present study, the findings revealed that the expression of YTHDF2 was positively correlated with MM stage, recurrence and resistance. Moreover, high expression of YTHDF2 was shown to be an independent and adverse factor for the OS of MM patients. In addition, we showed that silencing YTHDF2 could not only inhibit MM proliferation but also cause the G1/S phase arrest in cell cycle. Similarly, Huang et al. [38] reported that YTHDF2 was upregulated in intrahepatic cholangiocarcinoma, and its knockdown resulted in the suppressed proliferation and G0/G1 cell cycle arrest by promoting CDKN1B (p27) degradation, further validating the role of YTHDF2 in cell cycle regulation.
To further demonstrate the potential mechanism through which YTHDF2 promoted MM proliferation and cell cycle regulation, we analyzed RNA-seq and CLIP-seq data from public databases and identified EGR1 as a potential target for YTHDF2. RIP-qPCR and MeRIP-qPCR results demonstrated that YTHDF2 could interact with the EGR1 mRNA to accelerate its degradation in an m6A-dependent manner. Concordantly, Liao et al. [38] showed that EGR1 mRNA was the target for METTL3 and YTHDF3. The EGR1 mRNA underwent m6A modifications through the action of METTL3, and YTHDF3 enhanced its RNA stability to upregulate the EGR1/Snail signaling pathway and support cancer metastasis in esophageal squamous cell carcinoma.
The zinc finger transcription factor EGR1 can regulate cell life and death. It consists of a DNA-binding domain, a transcriptional activation domain and an inhibitory domain [39]. However, the role of EGR1 varies in different cancers. As the name suggests, EGR1 can be activated by many growth and inflammatory factors via the MAPK signaling pathway to promote cell proliferation [40]. For example, Grotegut et al. [41] reported that the hepatocyte growth factor could induce cell migration and invasion via the MAPK/EGR1/Snail signaling pathway in liver cancer. In addition, Fahmy et al. [42] showed that EGR1 was crucial for supporting fibroblast growth factor-dependent tumor angiogenesis and tumor growth in breast cancer. EGR1 can also exert its pro-apoptotic and tumor-inhibiting function by transcriptionally activating tumor suppressor genes, such as TP53, p21cip1/waf1, PTEN and JUN [23, 43, 44]. Virolle et al. [43] demonstrated that EGR1 directly transactivated PTEN to enhance radiation or chemotherapy-induced apoptosis, and the loss of EGR1 conferred cancer cells with radiation and drug resistance. In MM, mutation of EGR1 gene was observed in both MM and MGUS [45, 46]. Moreover, Chen et al. [47] reported that EGR1 expression was decreased in MM compared to MGUS and SMM, and low expression of EGR1 was highly associated with poor prognosis, consistent with our results. They further showed that EGR1 was the direct target for JUN, and was critical in bortezomib-induced apoptosis. In this study, we noted a decrease in the levels of EGR1 during the process of MM development. Moreover, knockdown of EGR1 promoted MM cell proliferation and cell cycle transition by decreasing p21cip1/waf1 expression and increasing CDK2-Cyclin E1 expression. Additionally, knockdown of EGR1 significantly reversed the inhibited proliferation and G1/S phase arrest induced by silencing YTHDF2.
The underlying mechanism contributing to the high expression of YTHDF2 in MM appears to be complex and remains unclear. Transcriptionally, the hypoxia-induced HIF1α/2α signaling pathway can negatively regulate YTHDF2 expression to favor tumor growth in liver cancer [48, 49]. On the other hand, hypoxia was reported to positively regulate the expression of YTHDF2 in lung cancer [50]. At the post-translational level, FBW7 was shown to be involved in ubiquitin-proteasome degradation of YTHDF2 protein in ovarian cancer [51]. In addition, the EGFR/Src/ERK cascade could positively mediate YTHDF2 expressing by stabilizing the YTHDF2 protein through phosphorylation of its serine 39 and threonine 381 [16]. Yang et al. recently reported that the interaction between MM cells and bone marrow adipocytes could promoted the release of adipocyte-derived exocomal LncRNA through the action of METTL17A-mediated m6A methylation [52], which further exacerbated MM therapeutic resistance, suggesting that the cellular interaction of MM cells in the bone marrow microenvironment can influence cellular m6A levels.
In conclusion, our study demonstrated that YTHDF2 was an independent prognostic factor in MM that could promote tumor growth and cell cycle transition via the EGR1/p21cip1/waf1/CDK2-Cyclin E1 axis, highlighting that YTHDF2 could be used as a potential biomarker for predicting prognosis and a promising therapeutic target in MM.
Data availability
The datasets used in this article were obtain from the public GEO database.
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA: A Cancer J Clin. 2022;72:7–33.
Zhao WH, Wang BY, Chen LJ, Fu WJ, Xu J, Liu J, et al. Four-year follow-up of LCAR-B38M in relapsed or refractory multiple myeloma: a phase 1, single-arm, open-label, multicenter study in China (LEGEND-2). J Hematol Oncol. 2022;15:86.
Adams JM, Cory S. Modified nucleosides and bizarre 5’-termini in mouse myeloma mRNA. Nature. 1975;255:28–33.
Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. 1974;71:3971–5.
Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–d307.
Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, et al. A majority of m6A residues are in the last exons, allowing the potential for 3’ UTR regulation. Genes Dev. 2015;29:2037–53.
Carroll SM, Narayan P, Rottman FM. N6-methyladenosine residues in an intron-specific region of prolactin pre-mRNA. Mol Cell Biol. 1990;10:4456–65.
Chen Z, Qi M, Shen B, Luo G, Wu Y, Li J, et al. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 2019;47:2533–45.
Wang X, Ma R, Zhang X, Cui L, Ding Y, Shi W, et al. Crosstalk between N6-methyladenosine modification and circular RNAs: current understanding and future directions. Mol Cancer. 2021;20:121.
Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18:103.
Watatani Y, Sato Y, Miyoshi H, Sakamoto K, Nishida K, Gion Y, et al. Molecular heterogeneity in peripheral T-cell lymphoma, not otherwise specified revealed by comprehensive genetic profiling. Leukemia. 2019;33:2867–83.
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–20.
Liu R, Jia Y, Kong G, He A. Novel insights into roles of N6-methyladenosine reader YTHDF2 in cancer progression. J Cancer Res Clin Oncol. 2022.
Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, et al. Targeting the RNA m(6)A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell. 2019;25:137–48.e136.
Li J, Xie H, Ying Y, Chen H, Yan H, He L, et al. YTHDF2 mediates the mRNA degradation of the tumor suppressors to induce AKT phosphorylation in N6-methyladenosine-dependent way in prostate cancer. Mol Cancer. 2020;19:152.
Fang R, Chen X, Zhang S, Shi H, Ye Y, Shi H, et al. EGFR/SRC/ERK-stabilized YTHDF2 promotes cholesterol dysregulation and invasive growth of glioblastoma. Nat Commun. 2021;12:177.
Huang CS, Chu J, Zhu XX, Li JH, Huang XT, Cai JP, et al. The C/EBPβ-LINC01133 axis promotes cell proliferation in pancreatic ductal adenocarcinoma through upregulation of CCNG1. Cancer Lett. 2018;421:63–72.
Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I, Lemma RB, Turchi L, Blanc-Mathieu R, et al. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022;50:D165–d173.
Zhao W, Zhang S, Zhu Y, Xi X, Bao P, Ma Z, et al. POSTAR3: an updated platform for exploring post-transcriptional regulation coordinated by RNA-binding proteins. Nucleic Acids Res. 2022;50:D287–d294.
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523.
Zhou Y, Zeng P, Li YH, Zhang Z, Cui Q. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016;44:e91.
Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–14.
Shin SY, Kim CG, Kim SH, Kim YS, Lim Y, Lee YH. Chlorpromazine activates p21Waf1/Cip1 gene transcription via early growth response-1 (Egr-1) in C6 glioma cells. Exp Mol Med. 2010;42:395–405.
Choi BH, Kim CG, Bae YS, Lim Y, Lee YH, Shin SY. p21 Waf1/Cip1 expression by curcumin in U-87MG human glioma cells: role of early growth response-1 expression. Cancer Res. 2008;68:1369–77.
Kim CG, Choi BH, Son SW, Yi SJ, Shin SY, Lee YH. Tamoxifen-induced activation of p21Waf1/Cip1 gene transcription is mediated by Early Growth Response-1 protein through the JNK and p38 MAP kinase/Elk-1 cascades in MDA-MB-361 breast carcinoma cells. Cell Signal. 2007;19:1290–300.
Schmidt K, Carroll JS, Yee E, Thomas DD, Wert-Lamas L, Neier SC, et al. The lncRNA SLNCR Recruits the Androgen Receptor to EGR1-Bound Genes in Melanoma and Inhibits Expression of Tumor Suppressor p21. Cell Rep. 2019;27:2493–507.e2494.
Kim SJ, Kim JM, Shim SH, Chang HI. Shikonin induces cell cycle arrest in human gastric cancer (AGS) by early growth response 1 (Egr1)-mediated p21 gene expression. J Ethnopharmacol. 2014;151:1064–71.
Kumar SK, Rajkumar SV. The multiple myelomas - current concepts in cytogenetic classification and therapy. Nat Rev Clin Oncol. 2018;15:409–21.
Cardona-Benavides IJ, de Ramón C, Gutiérrez NC. Genetic Abnormalities in Multiple Myeloma: Prognostic and Therapeutic Implications. Cells. 2021;10:336.
Fong CY, Morison J, Dawson MA. Epigenetics in the hematologic malignancies. Haematologica. 2014;99:1772–83.
Zhu T, Roundtree IA, Wang P, Wang X, Wang L, Sun C, et al. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 2014;24:1493–6.
Li F, Zhao D, Wu J, Shi Y. Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition. Cell Res. 2014;24:1490–2.
Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626.
Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, et al. Endoribonucleolytic Cleavage of m(6)A-Containing RNAs by RNase P/MRP Complex. Mol Cell. 2019;74:494–507.e498.
Li Y, Sheng H, Ma F, Wu Q, Huang J, Chen Q, et al. RNA m(6)A reader YTHDF2 facilitates lung adenocarcinoma cell proliferation and metastasis by targeting the AXIN1/Wnt/β-catenin signaling. Cell Death Dis. 2021;12:479.
Zhong L, Liao D, Zhang M, Zeng C, Li X, Zhang R, et al. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252–61.
Hua Z, Wei R, Guo M, Lin Z, Yu X, Li X, et al. YTHDF2 promotes multiple myeloma cell proliferation via STAT5A/MAP2K2/p-ERK axis. Oncogene. 2022;41:1482–91.
Huang CS, Zhu YQ, Xu QC, Chen S, Huang Y, Zhao G, et al. YTHDF2 promotes intrahepatic cholangiocarcinoma progression and desensitises cisplatin treatment by increasing CDKN1B mRNA degradation. Clin Transl Med. 2022;12:e848.
Sukhatme VP, Cao XM, Chang LC, Tsai-Morris CH, Stamenkovich D, Ferreira PC, et al. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell. 1988;53:37–43.
Thiel G, Cibelli G. Regulation of life and death by the zinc finger transcription factor Egr-1. J Cell Physiol. 2002;193:287–92.
Grotegut S, von Schweinitz D, Christofori G, Lehembre F. Hepatocyte growth factor induces cell scattering through MAPK/Egr-1-mediated upregulation of Snail. EMBO J. 2006;25:3534–45.
Fahmy RG, Dass CR, Sun LQ, Chesterman CN, Khachigian LM. Transcription factor Egr-1 supports FGF-dependent angiogenesis during neovascularization and tumor growth. Nat Med. 2003;9:1026–32.
Virolle T, Adamson ED, Baron V, Birle D, Mercola D, Mustelin T, et al. The Egr-1 transcription factor directly activates PTEN during irradiation-induced signalling. Nat Cell Biol. 2001;3:1124–8.
de Belle I, Huang RP, Fan Y, Liu C, Mercola D, Adamson ED. p53 and Egr-1 additively suppress transformed growth in HT1080 cells but Egr-1 counteracts p53-dependent apoptosis. Oncogene. 1999;18:3633–42.
Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997.
Mikulasova A, Wardell CP, Murison A, Boyle EM, Jackson GH, Smetana J, et al. The spectrum of somatic mutations in monoclonal gammopathy of undetermined significance indicates a less complex genomic landscape than that in multiple myeloma. Haematologica. 2017;102:1617–25.
Chen L, Wang S, Zhou Y, Wu X, Entin I, Epstein J, et al. Identification of early growth response protein 1 (EGR-1) as a novel target for JUN-induced apoptosis in multiple myeloma. Blood. 2010;115:61–70.
Hou J, Zhang H, Liu J, Zhao Z, Wang J, Lu Z, et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol Cancer. 2019;18:163.
Chen Z, Shao YL, Wang LL, Lin J, Zhang JB, Ding Y, et al. YTHDF2 is a potential target of AML1/ETO-HIF1α loop-mediated cell proliferation in t(8;21) AML. Oncogene. 2021;40:3786–98.
Xu P, Hu K, Zhang P, Sun ZG, Zhang N. Hypoxia-mediated YTHDF2 overexpression promotes lung squamous cell carcinoma progression by activation of the mTOR/AKT axis. Cancer Cell Int. 2022;22:13.
Xu F, Li J, Ni M, Cheng J, Zhao H, Wang S, et al. FBW7 suppresses ovarian cancer development by targeting the N(6)-methyladenosine binding protein YTHDF2. Mol Cancer. 2021;20:45.
Wang Z, He J, Bach DH, Huang YH, Li Z, Liu H, et al. Induction of m(6)A methylation in adipocyte exosomal LncRNAs mediates myeloma drug resistance. J Exp Clin Cancer Res: CR. 2022;41:4.
Funding
This work was supported by the Science and Technology Program of Shaanxi Province (Grant Number:2018ZDXM-SF-039).
Author information
Authors and Affiliations
Contributions
Conceptualization: ALH, RL, YY, GYK, JSH, JYM, FH; Methodology: RL, JYM, YCJ, FML, MZ, RZ, FH, JXL, XZX, TW, HL; Data curation, formal analysis, software and original draft: RL, JYM, YCJ; Supervision: ALH, YY, JSH, JYM, FH; Writing-review and editing: ALH, YY, GYK, JSH, JYM, YCJ, FH; Project administration: ALH.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The clinical samples used in this study to identify the YTHDF2 expression level were supervised and granted by the Ethics Committee of the Second Affiliated Hospital of Xi’an Jiaotong University (2015186). The animal experiments were supervised and granted by the Ethic Committee of Xi’an Jiaotong University Health Science Center (2022-1497).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, R., Miao, J., Jia, Y. et al. N6-methyladenosine reader YTHDF2 promotes multiple myeloma cell proliferation through EGR1/p21cip1/waf1/CDK2-Cyclin E1 axis-mediated cell cycle transition. Oncogene 42, 1607–1619 (2023). https://doi.org/10.1038/s41388-023-02675-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-023-02675-w