Abstract
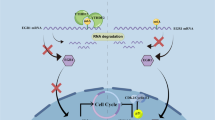
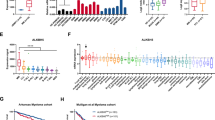
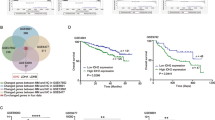
Multiple myeloma (MM) is still incurable partially due to lacking effective therapeutic targets. Aberrant N6-methyladenosine (m6A) RNA modification plays a vital role in many cancers, however few researches are executed in MM. We first screened the m6A-related genes in MM patient cohorts and correlated these genes with patient outcomes. We found that YTHDF2, a well-recognized m6A reader, was increased in MM patients and associated with poor outcomes. Decreased YTHDF2 expression hampered MM cell proliferation in vitro and in vivo, while enforced YTHDF2 expression reversed those effects. The analyses of m6A-RIP-seq and RIP-PCR indicated that STAT5A was the downstream target of YTHDF2, which was binding to the m6A modification site of STAT5A to promote its mRNA degradation. ChIP-seq and PCR assays revealed that STAT5A suppressed MM cell proliferation by occupying the transcription site of MAP2K2 to decrease ERK phosphorylation. In addition, we confirmed that YTHDF2 mediated the unphosphorylated form of STAT5A to inhibit the expression of MAP2K2/p-ERK. In conclusion, our study highlights that YTHDF2/STAT5A/MAP2K2/p-ERK axis plays a key role in MM proliferation and targeting YTHDF2 may be a promising therapeutic strategy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30.
Teras LR, DeSantis CE, Cerhan JR, Morton LM, Jemal A, Flowers CR. US lymphoid malignancy statistics by World Health Organization subtypes. CA Cancer J Clin. 2016;66:443–59.
Kumar SK, Rajkumar V, Kyle RA, van Duin M, Sonneveld P, Mateos MV, et al. Multiple myeloma. Nat Rev Dis Prim. 2017;3:17046.
Manasanch EE, Orlowski RZ. Proteasome inhibitors in cancer therapy. Nat Rev Clin Oncol. 2017;14:417–33.
Röllig C, Knop S, Bornhäuser M. Multiple myeloma. Lancet. 2015;385:2197–208.
Bianchi G, Anderson KC. Understanding biology to tackle the disease: multiple myeloma from bench to bedside, and back. CA Cancer J Clin. 2014;64:422–44.
Sonneveld P, Avet-Loiseau H, Lonial S, Usmani S, Siegel D, Anderson KC, et al. Treatment of multiple myeloma with high-risk cytogenetics: a consensus of the International Myeloma Working Group. Blood. 2016;127:2955–62.
Zhao Z, Chen Y, Francisco NM, Zhang Y, Wu M. The application of CAR-T cell therapy in hematological malignancies: advantages and challenges. Acta Pharm Sin B. 2018;8:539–51.
Rajkumar SV. Multiple myeloma: 2016 update on diagnosis, risk-stratification, and management. Am J Hematol. 2016;91:719–34.
Cardona-Benavides IJ, de Ramón C, Gutiérrez NC. Genetic abnormalities in multiple myeloma: prognostic and therapeutic implications. Cells. 2021;10:336.
Dimopoulos K, Gimsing P, Grønbæk K. The role of epigenetics in the biology of multiple myeloma. Blood Cancer J. 2014;4:e207.
Brioli A, Melchor L, Cavo M, Morgan GJ. The impact of intra-clonal heterogeneity on the treatment of multiple myeloma. Br J Haematol. 2014;165:441–54.
van Nieuwenhuijzen N, Spaan I, Raymakers R, Peperzak V. From MGUS to multiple myeloma, a paradigm for clonal evolution of premalignant cells. Cancer Res. 2018;78:2449–56.
He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18:176.
Wang T, Kong S, Tao M, Ju S. The potential role of RNA N6-methyladenosine in cancer progression. Mol Cancer. 2020;19:88.
Hu BB, Wang XY, Gu XY, Zou C, Gao ZJ, Zhang H, et al. N(6)-methyladenosine (m(6)A) RNA modification in gastrointestinal tract cancers: roles, mechanisms, and applications. Mol Cancer. 2019;18:178.
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–6.
Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′UTRs and near stop codons. Cell. 2012;149:1635–46.
Chen Y, Lin Y, Shu Y, He J, Gao W. Interaction between N(6)-methyladenosine (m(6)A) modification and noncoding RNAs in cancer. Mol Cancer. 2020;19:94.
Bach C, Leffler M, Kroenke J, Saul D, Mougiakakos D, Mackensen A, et al. Role of N6-methyladenosine (m6A) RNA modification in multiple myeloma. Oncol Res Treat. 2019;42:123.
Bai H, Xu P, Chen B. Gene signatures and prognostic values of m6A-related genes in multiple myeloma. Curr Res Transl Med. 2021;69:103288.
Jiang F, Tang X, Tang C, Hua Z, Ke M, Wang C, et al. HNRNPA2B1 promotes multiple myeloma progression by increasing AKT3 expression via m6A-dependent stabilization of ILF3 mRNA. J Hematol Oncol. 2021;14:54.
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–20.
Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626.
Shen X, Zhao K, Xu L, Cheng G, Zhu J, Gan L, et al. YTHDF2 inhibits gastric cancer cell growth by regulating FOXC2 signaling pathway. Front Genet. 2020;11:592042.
Sheng H, Li Z, Su S, Sun W, Zhang X, Li L, et al. YTH domain family 2 promotes lung cancer cell growth by facilitating 6-phosphogluconate dehydrogenase mRNA translation. Carcinogenesis. 2020;41:541–50.
Zhong L, Liao D, Zhang M, Zeng C, Li X, Zhang R, et al. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252–61.
Huang T, Liu Z, Zheng Y, Feng T, Gao Q, Zeng W. YTHDF2 promotes spermagonial adhesion through modulating MMPs decay via m(6)A/mRNA pathway. Cell Death Dis. 2020;11:37.
Neuzillet C, Tijeras-Raballand A, de Mestier L, Cros J, Faivre S, Raymond E. MEK in cancer and cancer therapy. Pharmacol Ther. 2014;141:160–71.
Hu X, Dutta P, Tsurumi A, Li J, Wang J, Land H, et al. Unphosphorylated STAT5A stabilizes heterochromatin and suppresses tumor growth. Proc Natl Acad Sci USA. 2013;110:10213–8.
Li WX. Canonical and non-canonical JAK-STAT signaling. Trends Cell Biol. 2008;18:545–51.
Liao W, Lin JX, Leonard WJ. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity. 2013;38:13–25.
Bakhoum SF, Landau DA. Chromosomal instability as a driver of tumor heterogeneity and evolution. Cold Spring Harb Perspect Med. 2017;7:a029611.
Neuse CJ, Lomas OC, Schliemann C, Shen YJ, Manier S, Bustoros M, et al. Genome instability in multiple myeloma. Leukemia. 2020;34:2887–97.
Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997.
Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25:91–101.
Robiou du Pont S, Cleynen A, Fontan C, Attal M, Munshi N, Corre J, et al. Genomics of multiple myeloma. J Clin Oncol. 2017;35:963–7.
Gu C, Wang W, Tang X, Xu T, Zhang Y, Guo M, et al. CHEK1 and circCHEK1_246aa evoke chromosomal instability and induce bone lesion formation in multiple myeloma. Mol Cancer. 2021;20:84.
Tang X, Guo M, Ding P, Deng Z, Ke M, Yuan Y, et al. BUB1B and circBUB1B_544aa aggravate multiple myeloma malignancy through evoking chromosomal instability. Signal Transduct Target Ther. 2021;6:361.
Zhu H, Jia X, Wang Y, Song Z, Wang N, Yang Y, et al. M6A classification combined with tumor microenvironment immune characteristics analysis of bladder cancer. Front Oncol. 2021;11:714267.
Shen X, Hu B, Xu J, Qin W, Fu Y, Wang S, et al. The m6A methylation landscape stratifies hepatocellular carcinoma into 3 subtypes with distinct metabolic characteristics. Cancer Biol Med. 2020;17:937–52.
McClure JJ, Li X, Chou CJ. Advances and challenges of HDAC inhibitors in cancer therapeutics. Adv Cancer Res. 2018;138:183–211.
Brown S, Pawlyn C, Tillotson AL, Sherratt D, Flanagan L, Low E, et al. Bortezomib, vorinostat, and dexamethasone combination therapy in relapsed myeloma: results of the phase 2 MUK four trial. Clin Lymphoma Myeloma Leuk. 2021;21:154–61.e3.
Chen B, Li Y, Song R, Xue C, Xu F. Functions of RNA N6-methyladenosine modification in cancer progression. Mol Biol Rep. 2019;46:2567–75.
Chang YZ, Chai RC, Pang B, Chang X, An SY, Zhang KN, et al. METTL3 enhances the stability of MALAT1 with the assistance of HuR via m6A modification and activates NF-κB to promote the malignant progression of IDH-wildtype glioma. Cancer Lett. 2021;511:36–46.
Liu Y, Liang G, Xu H, Dong W, Dong Z, Qiu Z, et al. Tumors exploit FTO-mediated regulation of glycolytic metabolism to evade immune surveillance. Cell Metab. 2021;33:1221–33.
Xu Y, Liu J, Chen WJ, Ye QQ, Chen WT, Li CL, et al. Regulation of N6-methyladenosine in the differentiation of cancer stem cells and their fate. Front Cell Dev Biol. 2020;8:561703.
Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother. 2019;112:108613.
Song P, Feng L, Li J, Dai D, Zhu L, Wang C, et al. β-catenin represses miR455-3p to stimulate m6A modification of HSF1 mRNA and promote its translation in colorectal cancer. Mol Cancer. 2020;19:129.
Zhang L, Wan Y, Zhang Z, Jiang Y, Lang J, Cheng W, et al. FTO demethylates m6A modifications in HOXB13 mRNA and promotes endometrial cancer metastasis by activating the WNT signalling pathway. RNA Biol. 2021;18:1265–78.
Cui X, Wang Z, Li J, Zhu J, Ren Z, Zhang D, et al. Cross talk between RNA N6-methyladenosine methyltransferase-like 3 and miR-186 regulates hepatoblastoma progression through Wnt/β-catenin signalling pathway. Cell Prolif. 2020;53:e12768.
Song S, Fan G, Li Q, Su Q, Zhang X, Xue X, et al. IDH2 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in multiple myeloma. Oncogene. 2021;40:5393–402.
Ri M. Endoplasmic-reticulum stress pathway-associated mechanisms of action of proteasome inhibitors in multiple myeloma. Int J Hematol. 2016;104:273–80.
Zhuang J, Shirazi F, Singh RK, Kuiatse I, Wang H, Lee HC, et al. Ubiquitin-activating enzyme inhibition induces an unfolded protein response and overcomes drug resistance in myeloma. Blood. 2019;133:1572–84.
Li Z, Qian P, Shao W, Shi H, He XC, Gogol M, et al. Suppression of m(6)A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res. 2018;28:904–17.
Xu J, Pfarr N, Endris V, Mai EK, Md Hanafiah NH, Lehners N, et al. Molecular signaling in multiple myeloma: association of RAS/RAF mutations and MEK/ERK pathway activation. Oncogenesis. 2017;6:e337.
Heuck CJ, Jethava Y, Khan R, van Rhee F, Zangari M, Chavan S, et al. Inhibiting MEK in MAPK pathway-activated myeloma. Leukemia. 2016;30:976–80.
Wu CJ, Sundararajan V, Sheu BC, Huang RY, Wei LH. Activation of STAT3 and STAT5 signaling in epithelial ovarian cancer progression: mechanism and therapeutic opportunity. Cancers. 2019;12:24.
Lauta VM. A review of the cytokine network in multiple myeloma: diagnostic, prognostic, and therapeutic implications. Cancer. 2003;97:2440–52.
Peng Y, Li F, Zhang P, Wang X, Shen Y, Feng Y, et al. IGF-1 promotes multiple myeloma progression through PI3K/Akt-mediated epithelial-mesenchymal transition. Life Sci. 2020;249:117503.
Furigo IC, Melo HM. Lyra ESNM, Ramos-Lobo AM, Teixeira PDS, Buonfiglio DC, et al. Brain STAT5 signaling modulates learning and memory formation. Brain Struct Funct. 2018;223:2229–41.
Kimura A, Martin C, Robinson GW, Simone JM, Chen W, Wickre MC, et al. The gene encoding the hematopoietic stem cell regulator CCN3/NOV is under direct cytokine control through the transcription factors STAT5A/B. J Biol Chem. 2010;285:32704–9.
Rani A, Murphy JJ. STAT5 in cancer and immunity. J Interferon Cytokine Res. 2016;36:226–37.
Cimino G, Avvisati G, Amadori S, Cava MC, Giannarelli D, Di Nucci GD, et al. High serum IL-2 levels are predictive of prolonged survival in multiple myeloma. Br J Haematol. 1990;75:373–7.
Surbek M, Tse W, Moriggl R, Han X. A centric view of JAK/STAT5 in intestinal homeostasis, infection, and inflammation. Cytokine. 2021;139:155392.
Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, et al. The molecular classification of multiple myeloma. Blood. 2006;108:2020–8.
Broyl A, Hose D, Lokhorst H, de Knegt Y, Peeters J, Jauch A, et al. Gene expression profiling for molecular classification of multiple myeloma in newly diagnosed patients. Blood. 2010;116:2543–53.
Wei R, Zhong S, Qiao L, Guo M, Shao M, Wang S, et al. Steroid 5α-reductase type I induces cell viability and migration via nuclear factor-κB/vascular endothelial growth factor signaling pathway in colorectal cancer. Front Oncol. 2020;10:1501.
Acknowledgements
This work was supported by National Natural Science Foundation of China 81970196 (to CG), 82073885 (to YY), 82003832 (to MG); Natural Science Foundation of Jiangsu Province BK20200097 (to CG); Jiangsu Postgraduate Research and Practice Innovation Program KYCX21_1769 (to RW); A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (Integration of Chinese and Western Medicine).
Author information
Authors and Affiliations
Contributions
YY and CG designed and conceived the experiments. ZH and RW developed methodology and conducted most of the experiments. MG analyzed and processed the sequencing data. ZL performed the animal experiments. XY and XL acquired the data. ZH and CG drafted the manuscript. YY and CG conducted supervision and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Hua, Z., Wei, R., Guo, M. et al. YTHDF2 promotes multiple myeloma cell proliferation via STAT5A/MAP2K2/p-ERK axis. Oncogene 41, 1482–1491 (2022). https://doi.org/10.1038/s41388-022-02191-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02191-3
This article is cited by
-
m6A reader IGF2BP2 promotes lymphatic metastasis by stabilizing DPP4 in papillary thyroid carcinoma
Cancer Gene Therapy (2024)
-
circSKA3 promotes colorectal cancer metastases through miR-1238 and methylation
Molecular and Cellular Biochemistry (2024)
-
N6-methyladenosine reader YTHDF2 promotes multiple myeloma cell proliferation through EGR1/p21cip1/waf1/CDK2-Cyclin E1 axis-mediated cell cycle transition
Oncogene (2023)
-
PRMT1 methylation of WTAP promotes multiple myeloma tumorigenesis by activating oxidative phosphorylation via m6A modification of NDUFS6
Cell Death & Disease (2023)
-
Novel insights into roles of N6-methyladenosine reader YTHDF2 in cancer progression
Journal of Cancer Research and Clinical Oncology (2022)