Abstract
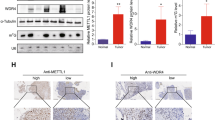
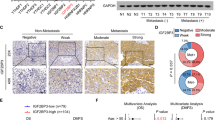
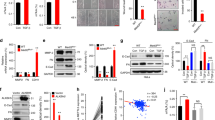
Treatment selections are very limited for patients with advanced nasopharyngeal carcinoma (NPC) experiencing disease progression. Uncovering mechanisms underlying NPC progression is crucial for the development of novel treatments. Here we show that N7-methylguanosine (m7G) tRNA modification enzyme METTL1 and its partner WDR4 are significantly elevated in NPC and are associated with poor prognosis. Loss-of-function and gain-of-function assays demonstrated that METTL1/WDR4 promotes NPC growth and metastasis in vitro and in vivo. Mechanistically, ARNT was identified as an upstream transcription factor regulating METTL1 expression in NPC. METTL1 depletion resulted in decreased m7G tRNA modification and expression, which led to impaired codon recognition during mRNA translation, therefore reducing the translation efficiencies of mRNAs with higher m7G codons. METTL1 upregulated the WNT/β-catenin signaling pathway and promoted NPC cell epithelial-mesenchymal transition (EMT) and chemoresistance to cisplatin and docetaxel in vitro and in vivo. Overexpression of WNT3A bypassed the requirement of METTL1 for EMT and chemoresistance. This work uncovers novel insights into tRNA modification-mediated mRNA translation regulation and highlights the critical function of tRNA modification in cancer progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data and code availability
The previously published Microarray data that were re-analyzed here are available under the accession code GEO12452 and GEO103611. The chemicals and commercial kits used in this study were shown in Table S7. The polyribosome-RNA-seq, Ribo-seq and m7G tRNA TRAC-seq data generated in this study are available at NCBI GEO DataSet (GSE169589). Software and algorithms employed in this study were presented in Supplementary Table S8.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Wu LR, Liu YT, Jiang N, Fan YX, Wen J, Huang SF, et al. Ten-year survival outcomes for patients with nasopharyngeal carcinoma receiving intensity-modulated radiotherapy: An analysis of 614 patients from a single center. Oral Oncol. 2017;69:26–32.
Mao YP, Xie FY, Liu LZ, Sun Y, Li L, Tang LL, et al. Re-evaluation of 6th edition of AJCC staging system for nasopharyngeal carcinoma and proposed improvement based on magnetic resonance imaging. Int J Radiat Oncol Biol Phys. 2009;73:1326–34.
Pan JJ, Ng WT, Zong JF, Chan LL, O’Sullivan B, Lin SJ, et al. Proposal for the 8th edition of the AJCC/UICC staging system for nasopharyngeal cancer in the era of intensity-modulated radiotherapy. Cancer 2016;122:546–58.
Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. Science 2018;361:1346–9.
Han X, Wang M, Zhao YL, Yang Y, Yang YG. RNA methylations in human cancers. Semin Cancer Biol. 2020;18:S1044-579X(20)30241-8.
Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nuclic Acids Res. 2018;46:D303–D7.
Boccaletto P, Magnus M, Almeida C, Zyla A, Astha A, Pluta R, et al. RNArchitecture: A database and a classification system of RNA families, with a focus on structural information. Nuclic Acids Res. 2018;46:D202–D5.
Pinello N, Sun S, Wong JJ. Aberrant expression of enzymes regulating m(6)A mRNA methylation: Implication in cancer. Cancer Biol Med. 2018;15:323–34.
Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18:103.
Huo FC, Zhu ZM, Pei DS. N(6) -methyladenosine (m(6) A) RNA modification in human cancer. Cell Prolif. 2020;53:e12921.
Liu J, Harada BT, He C. Regulation of Gene Expression by N(6)-methyladenosine in Cancer. Trends Cell Biol. 2019;29:487–99.
Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat Rev Mol Cell Biol. 2018;19:45–58.
Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, et al. ALKBH1-Mediated tRNA Demethylation Regulates Translation. Cell 2016;167:1897.
Chou HJ, Donnard E, Gustafsson HT, Garber M, Rando OJ. Transcriptome-wide Analysis of Roles for tRNA Modifications in Translational Regulation. Mol Cell. 2017;68:978–92 e4.
Torres AG, Batlle E, Ribas de Pouplana L. Role of tRNA modifications in human diseases. Trends Mol Med. 2014;20:306–14.
Kirchner S, Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet. 2015;16:98–112.
Lin S, Liu Q, Lelyveld VS, Choe J, Szostak JW, Gregory RI. Mettl1/Wdr4-Mediated m(7)G tRNA Methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol Cell. 2018;71:244–55 e5.
Alexandrov A, Martzen MR, Phizicky EM. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 2002;8:1253–66.
Trimouille A, Lasseaux E, Barat P, Deiller C, Drunat S, Rooryck C, et al. Further delineation of the phenotype caused by biallelic variants in the WDR4 gene. Clin Genet. 2018;93:374–7.
Chen X, Gao Y, Yang L, Wu B, Dong X, Liu B, et al. Speech and language delay in a patient with WDR4 mutations. Eur J Med Genet. 2018;61:468–72.
Shaheen R, Abdel-Salam GM, Guy MP, Alomar R, Abdel-Hamid MS, Afifi HH, et al. Mutation in WDR4 impairs tRNA m(7)G46 methylation and causes a distinct form of microcephalic primordial dwarfism. Genome Biol. 2015;16:210.
Dai Z, Liu H, Liao J, Huang C, Ren X, Zhu W, et al. N(7)-Methylguanosine tRNA modification enhances oncogenic mRNA translation and promotes intrahepatic cholangiocarcinoma progression. Mol Cell. 2021;81:3339–55.
Ma J, Han H, Huang Y, Yang C, Zheng S, Cai T, et al. METTL1/WDR4-mediated m(7)G tRNA modifications and m(7)G codon usage promote mRNA translation and lung cancer progression. Mol Ther. 2021;29:3422–35.
Orellana EA, Liu Q, Yankova E, Pirouz M, De Braekeleer E, Zhang W, et al. METTL1-mediated m(7)G modification of Arg-TCT tRNA drives oncogenic transformation. Mol Cell. 2021;81:3323–38.
Mao YP, Liang SB, Liu LZ, Chen Y, Sun Y, Tang LL, et al. The N staging system in nasopharyngeal carcinoma with radiation therapy oncology group guidelines for lymph node levels based on magnetic resonance imaging. Clin Cancer Res. 2008;14:7497–503.
Terashima J, Sampei S, Iidzuka M, Ohsakama A, Tachikawa C, Satoh J, et al. VEGF expression is regulated by HIF-1alpha and ARNT in 3D KYSE-70, esophageal cancer cell spheroids. Cell Biol Int. 2016;40:1187–94.
Leick KM, Obeid JM, Bekiranov S, Slingluff CL Jr. Systems analysis of barrier molecule and ARNT-related gene expression regulation in melanoma. Oncoimmunology 2019;8:e1665978.
Chen G, Cai ZD, Lin ZY, Wang C, Liang YX, Han ZD, et al. ARNT-dependent CCR8 reprogrammed LDH isoform expression correlates with poor clinical outcomes of prostate cancer. Mol Carcinog. 2020;59:897–907.
Persson CU, von Stedingk K, Fredlund E, Bexell D, Pahlman S, Wigerup C, et al. ARNT-dependent HIF-2 transcriptional activity is not sufficient to regulate downstream target genes in neuroblastoma. Exp Cell Res. 2020;388:111845.
Lin S, Liu Q, Jiang YZ, Gregory RI. Nucleotide resolution profiling of m(7)G tRNA modification by TRAC-Seq. Nat Protoc. 2009;4:44–57.
Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6:275–7.
Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature 2001;411:349–54.
Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science 2004;303:1483–7.
Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–51.
Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–73.
Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015;527:472–6.
Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015;527:525–30.
Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7:re8.
Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96.
Bernaudo S, Salem M, Qi X, Zhou W, Zhang C, Yang W, et al. Cyclin G2 inhibits epithelial-to-mesenchymal transition by disrupting Wnt/beta-catenin signalling. Oncogene 2016;35:4828.
Ren X, Yang X, Cheng B, Chen X, Zhang T, He Q, et al. HOPX hypermethylation promotes metastasis via activating SNAIL transcription in nasopharyngeal carcinoma. Nat Commun. 2017;8:14053.
Zhang J, Xie T, Zhong X, Jiang HL, Li R, Wang BY, et al. Melatonin reverses nasopharyngeal carcinoma cisplatin chemoresistance by inhibiting the Wnt/beta-catenin signaling pathway. Aging 2020;12:5423–38.
Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62:335–45.
Zhang P, He Q, Lei Y, Li Y, Wen X, Hong M, et al. m(6)A-mediated ZNF750 repression facilitates nasopharyngeal carcinoma progression. Cell Death Dis. 2018;9:1169.
Chang G, Shi L, Ye Y, Shi H, Zeng L, Tiwary S, et al. YTHDF3 induces the translation of m(6)A-enriched gene transcripts to promote breast cancer brain metastasis. Cancer Cell. 2020;14:857–71.e7.
Liu L, Wu Y, Li Q, Liang J, He Q, Zhao L, et al. METTL3 promotes tumorigenesis and metastasis through BMI1 m(6)A methylation in oral squamous cell carcinoma. Mol Ther. 2020;28:2177–90.
Shen C, Xuan B, Yan T, Ma Y, Xu P, Tian X, et al. m(6)A-dependent glycolysis enhances colorectal cancer progression. Mol Cancer. 2020;19:72.
Song P, Feng L, Li J, Dai D, Zhu L, Wang C, et al. beta-catenin represses miR455-3p to stimulate m6A modification of HSF1 mRNA and promote its translation in colorectal cancer. Mol Cancer. 2020;19:129.
Pan T. Modifications and functional genomics of human transfer RNA. Cell Res. 2018;28:395–404.
Michaud J, Kudoh J, Berry A, Bonne-Tamir B, Lalioti MD, Rossier C, et al. Isolation and characterization of a human chromosome 21q22.3 gene (WDR4) and its mouse homologue that code for a WD-repeat protein. Genomics 2000;68:71–9.
Najmabadi H, Hu H, Garshasbi M, Zemojtel T, Abedini SS, Chen W, et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 2011;478:57–63.
Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020–39.
Guy MP, Shaw M, Weiner CL, Hobson L, Stark Z, Rose K, et al. Defects in tRNA anticodon Loop 2’-O-Methylation are implicated in nonsyndromic X-linked intellectual disability due to mutations in FTSJ1. Hum Mutat. 2015;36:1176–87.
Shaheen R, Han L, Faqeih E, Ewida N, Alobeid E, Phizicky EM, et al. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum Genet. 2016;135:707–13.
Tian QH, Zhang MF, Zeng JS, Luo RG, Wen Y, Chen J, et al. METTL1 overexpression is correlated with poor prognosis and promotes hepatocellular carcinoma via PTEN. J Mol Med. 2019;97:1535–45.
Liu Y, Zhang Y, Chi Q, Wang Z, Sun B. Methyltransferase-like 1 (METTL1) served as a tumor suppressor in colon cancer by activating 7-methyguanosine (m7G) regulated let-7e miRNA/HMGA2 axis. Life Sci. 2020;249:117480.
Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet 2019;394:64–80.
Okamoto M, Fujiwara M, Hori M, Okada K, Yazama F, Konishi H, et al. tRNA modifying enzymes, NSUN2 and METTL1, determine sensitivity to 5-fluorouracil in HeLa cells. PLoS Genet. 2014;10:e1004639.
Kim SW, Li Z, Moore PS, Monaghan AP, Chang Y, Nichols M, et al. A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic Acids Res. 2010;38:e98.
Peng H, Zhang J, Zhang PP, Chen L, Tang LL, Yang XJ, et al. ARNTL hypermethylation promotes tumorigenesis and inhibits cisplatin sensitivity by activating CDK5 transcription in nasopharyngeal carcinoma. J Exp Clin Cancer Res. 2019;38:11.
Hong X, Liu N, Liang Y, He Q, Yang X, Lei Y, et al. Circular RNA CRIM1 functions as a ceRNA to promote nasopharyngeal carcinoma metastasis and docetaxel chemoresistance through upregulating FOXQ1. Mol Cancer. 2020;19:33.
Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13.
Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
Acknowledgements
We sincerely thank professor Jun Ma (Department of Radiation Oncology, Sun Yat-sen University Cancer Center) for kindly donate the NPC cell lines and nasopharyngeal normal cell lines. We sincerely thank Drs. Weidong Ji and Shuibin Lin for kindly sharing us with the plasmids for METTL1 overexpression and depletion. We thank all the patients who generously donate the tissues. The work was funded by National Natural Science Foundation of China (82002981, 82003232 and 82003212) and China Postdoctoral Science Foundation (2020M673003).
Author information
Authors and Affiliations
Contributions
BC and HP designed the protocol of this study. BC, JZ, PY and HP performed in vitro and in vivo assays. WJ and YH were responsible for the clinical sample tissues. BC performed TRAC-seq and data was analyzed by H.P. Polysome associated mRNA-seq and data analysis were performed by HP and PY. All other data analysis was performed by LW, HP and PY. BC and HP wrote the paper and all authors revised and modified the final paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Chen, B., Jiang, W., Huang, Y. et al. N7-methylguanosine tRNA modification promotes tumorigenesis and chemoresistance through WNT/β-catenin pathway in nasopharyngeal carcinoma. Oncogene 41, 2239–2253 (2022). https://doi.org/10.1038/s41388-022-02250-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02250-9
This article is cited by
-
PICK1 inhibits the malignancy of nasopharyngeal carcinoma and serves as a novel prognostic marker
Cell Death & Disease (2024)
-
Wnt/β-catenin-driven EMT regulation in human cancers
Cellular and Molecular Life Sciences (2024)
-
P300/SP1 complex mediating elevated METTL1 regulates CDK14 mRNA stability via internal m7G modification in CRPC
Journal of Experimental & Clinical Cancer Research (2023)
-
Biological roles of RNA m7G modification and its implications in cancer
Biology Direct (2023)
-
An innovative model based on N7-methylguanosine-related lncRNAs for forecasting prognosis and tumor immune landscape in bladder cancer
Cancer Cell International (2023)