Abstract
Exosomal microRNAs (miRNAs) have been suggested to participate in the pathogenesis of neuropsychiatric diseases, but their role in major depressive disorder (MDD) is unknown. We performed a genome-wide miRNA expression profiling of blood-derived exosomes from MDD patients and control subjects and revealed the top differentially expressed exosomal miRNA, i.e. hsa-miR-139-5p (upregulation), had good performance to differentiate between MDD patients and controls. Tail vein injection of blood exosomes isolated from MDD patients into normal mice caused their depressive-like behaviors as determined by the forced swimming, tail suspension, and novelty suppressed feeding tests, and injection of blood exosomes isolated from healthy volunteers into unpredictable mild stress (CUMS)-treated mice alleviated their depressive-like behaviors. CUMS mice also showed significantly increased blood and brain levels of exosomal miR-139-5p. Furthermore, the depressive-like behaviors in CUMS-treated mice were rescued by intranasal injection of miR-139-5p antagomir, suggesting that increased exosomal miR-139-5p levels may mediate stress-induced depression-like behavior in mice. Both exosome treatment and miR-139-5p antagomir treatment increased hippocampal neurogenesis in the CUMS-treated mice, and treatment of exosome from MDD patients decreased hippocampal neurogenesis in the normal mice. The role of miR-139-5p in neurogenesis was validated by in vitro experiments, demonstrating that miR-139-5p is a negative regulator for neural stem cell proliferation and neuronal differentiation. Our findings together suggest that exosomes from patients with major depression caused depressive-like behaviors in mice with involvement of miR-139-5p-regulated neurogenesis. Therefore, exosomal miRNAs are promising targets for the diagnosis and treatment of MDD.
Similar content being viewed by others
Introduction
Major depressive disorder (MDD) is one of the most common medical conditions leading to a significant increase in morbidity and mortality [1]. Despite considerable efforts in the field, our understanding of MDD pathophysiology remains rudimentary, and there are no validated biomarkers for this condition. Previous studies have focused on the role of monoamines including norepinephrine, dopamine, and serotonin in the pathogenesis of depression [2], and the first line of pharmacological treatment for MDD is selective serotonin reuptake inhibitors (SSRI) [3], and roughly half of MDD patients do not respond to SSRI treatment [4]. Recently, intranasal delivery of esketamine has been approved by FDA for the treatment of depression [5], although the use of ketamine in the patients with depression was reported to have various side effects [6] and over one third of depressed patients do not respond to ketamine [7]. Therefore, it is important to study the mechanism of MDD in order to identify MDD biomarkers and to develop new effective treatment.
Even though the cellular and molecular basis of neuropsychiatric diseases are still far from being understood, it is considered that cellular network disruption and aberrant information processing in the neural circuits play crucial roles in disease development [8]. In this context, exosomes, small vesicles 40–100 nm in size that are released by many cell types including neurons and glial cells, are gaining increasing attention due to their role in neurological disorders [9]. Exosomes contain a variety of molecules including DNAs, mRNAs, microRNAs (miRNAs), and proteins, which can be delivered to neighbor and distant cells through circulation, causing phenotypic changes in the recipient cell [9]. The transfer of exosomal content, especially miRNAs, has been implicated in various pathological processes with extensive studies in tumor metastasis formation [10]. Since synaptic plasticity has been found to regulate neuronal exosome release [11] and exosomes are able to cross the blood−brain barrier easily [12], circulating exosomes were suggested to be a valuable model for understanding the etiology of neuropsychiatric diseases [9, 11]. We recently showed evidence suggesting that dysregulation of blood exosome-derived miRNAs was not only involved in the pathogenesis of schizophrenia but also used as tools for schizophrenia diagnosis [13]. However, the potential role of exosomes in MDD pathogenesis has not been studied.
In the present study, we first explored serum exosome miRNA expression profile in patients with MDD and healthy control subjects to identify potential MDD markers by microRNA sequencing. Then, we studied blood and brain exosome-derived miRNA in an animal model of depression using mice subjected to chronic unpredictable mild stress (CUMS). Next, we manipulated blood exosomes and exosome-derived miRNA levels in mice and/or cell culture to explore the contribution of exosomes and exosome-derived miRNAs to depression development.
Materials and methods
Subjects and sample preparation
We recruited 33 drug-free MDD patients and 36 drug-free schizophrenia patients from the Third Hospital of Foshan, Foshan, China. The 46 healthy control subjects were recruited through advertisements. MDD and schizophrenia patients were diagnosed by experienced psychiatrists following the Structured Clinical Interview for DSM-IV (SCID) and International Classification of Diseases 10 (ICD-10). The Hamilton Depression Rating Scale (HAMD) and Montgomery-Asberg Depression Rating Scale (MADRS) were used to assess disease severity in MDD patients. Supplementary Table 1 shows the demographic and clinical variables of the participants.
All participants gave written informed consent before inclusion in the study. The study protocol was approved by the Ethics Committee at The Third People’s Hospital of Foshan, Foshan, China, and the experiments were conducted following the Declaration of Helsinki.
Blood exosome isolation and validation, and miRNA library construction and sequencing were performed as described previously [13], and brain exosome isolation was performed according to Vella et al. [14].
Differential expression and bioinformatic analysis
The duplicated miRNAs and miRNAs with a mean expression <10 transcripts per million (TPM) were removed. The remaining miRNAs were chosen to perform the differential expression analysis by DESeq2 [15] between MDD patients and healthy controls. False discovery rate (FDR) < 0.05 (corrected p value, Q value) was considered statistically significant for differentially expressed (DE) miRNA. The bioinformatics analyses were performed as previously described [13].
RNA extraction and qRT-PCR
RNA was extracted from blood exosomes (human or mouse) or whole brain using Trizol reagent (ThermoFisher, Waltham, MA, USA) and converted to cDNA using TaqManTM MicroRNA Reverse Transcription kit (ThermoFisher) following the manufacturer’s protocol. The levels of miR-139-5p were quantified using Taqman® MicroRNA Assay (including primers and probes, ThermoFisher) and normalized to the expression of miR-16. All qRT-PCR reactions were performed using a LightCycler® 96 Real-Time PCR Detection System (Roche, Basel, Switzerland).
Animals and treatments
Male C57BL/6 mice were obtained from Vital River Laboratory (Beijing, China). Animals were kept in a humidity- and temperature-controlled room on a 14:10 h light:dark cycle and given food and water ad libitum. Animal procedures were approved by the Animal Care and Use Committee of the Minzu University of China. The CUMS paradigm was established as described previously [16]. CUMS-treated mice and control mice were subjected to tail vein injection of exosome or saline, the exosomes were extracted from healthy volunteers and MDD patients and quantified using a nanoparticle tracking device ZetaView (Particle Metrix, Meerbusch, Germany) as described previously [13]. Each mouse received 1.4 × 1011 nanoparticles every 3 days, there were four injections in total.
For miR-139-5p antagomir experiments, the CUMS-treated mice and control mice were intranasally administered with 5 nmol miR-139-5p antagomir or negative control (NC) antagomir every 3 days for four times, and 2 mg of 5-BrdU (MedChemExpress, Monmouth Junction, New Jersey, USA) was intraperitoneally injected into the mice every day for 3 days before sacrifice to analyze newborn neurons.
Behavioral tests
After exosome or miR-139-5p antagomir treatments, we carried out the open-field, forced swimming, and tail suspension tests as described previously [16]. For the novelty suppressed feeding test, we measured the latency of the animals to approach and eat familiar food in a novel environment. The mice were food-deprived for 24 h before testing, a food pellet was placed on a white filter paper located in the middle of a test arena (55 × 55 × 40 cm), the mice were placed at one corner of the arena and allowed to explore for a maximum of 6 min. The latency to visit and begin chewing the food pellet was recorded.
Oxidative stress marker level/activity measurement
Serum samples were extracted from the mice after exosome treatments, and the samples were used to investigate oxidative stress status. Total antioxidant capacity (T-AOC), glutathione peroxidase (GSH-PX) activities and malondialdehyde (MDA) levels were measured by kits purchased from Nanjing Jiancheng (Nanjing, China).
Immunohistochemistry
To analyze adult hippocampal neurogenesis and/or newborn mature neurons after different treatments in the mice, we immunostained doublecortin (DCX), 5-BrdU and NeuN in the dentate gyrus of hippocampus by immunohistochemistry as described previously [17]. Polyclonal rabbit anti-DCX antibody (1:2000) was from Cell Signaling Technology (CST, Boston, MA, USA), monoclonal rat anti-BrdU antibody (1:800) was from Abcam (Cambridge, UK) and monoclonal mouse anti-NeuN antibody (1:800) was from Millipore (Burlington, Massachusetts, USA). The secondary antibodies were Alexa Fluor 594 goat anti-rabbit secondary antibody (1:1000, Invitrogen, Carlsbad, CA), Alexa Fluor 488 goat anti-mouse secondary antibody (1:1000, Invitrogen) and Alexa Fluor 594 goat anti-rat secondary antibody (1:1000, Invitrogen).
Proteomics
The mouse hippocampus was extracted and digested by the filter-aided sample preparation method [18], analyzed by the liquid chromatography-tandem mass spectrometry (LC-MS/MS) data-independent acquisition method, and quantified using Skyline [19] with the UniProt mouse protein database (see the supplementary methods for details).
Neurosphere culture preparation, transfection, proliferation and differentiation assays
Primary NSCs were obtained from E13.5 rats as described previously [20]. For the cell proliferation analysis, neurospheres were digested into single-cell suspensions, seeded in 96-well plates, and grown for 2 days to obtain neurosphere clones. Then, NSCs were transfected with 100 nM or 10 nM miR-139-5p mimic (miR-139-5p), miRNA scrambled control (miR-control), 50 nM miR-139-5p inhibitor (anti-miR-139-5p), and inhibitor scrambled control (anti-miR-control) according to the manufacturer’s instructions (RiboBio Co., Ltd., Guangzhou, China). The water-soluble tetrazolium-1 (WST-1) assay was performed to detect cell viability by measuring the optical density at 450 nm 6, 24, 48, 72, 96, and 120 h post transfection. For the neurosphere assay, images were taken by Leica DMi8 (Leica, Solms Germany) and the number and size of the neurospheres after 72 h transfection were calculated using ImageJ software.
For neuronal differentiation assay, neurosphere clones were transfected with miR-139-5p, miR-control, anti-139-5p and anti-miR-control, and then were allowed to differentiate for 120 h for immunocytochemistry.
Immunocytochemistry
Seventy-two hours (for proliferation) or 120 h (for differentiation) after transfection in NSCs, we performed immunocytochemistry in the cells using standard protocols [20] and examined the cells using a confocal microscope-Leica SP8 MP (Leica, Solms, Germany). Monoclonal rabbit anti-Sox2 antibody (1:1000), monoclonal mouse anti-glial fibrillary acidic protein (GFAP) antibody (1:1000) and monoclonal rabbit anti-microtubule-associated protein 2 (MAP2) antibody (1:1000) were from CST, and monoclonal mouse anti-Nestin antibody was from Proteintech (1:1000, Chicago, IL, USA). The secondary antibodies were Alexa Fluor 594 goat anti-rabbit secondary antibody (1:1000, Invitrogen) and Alexa Fluor 488 goat anti-mouse secondary antibody (1:1000, Invitrogen).
Western blot
One hundred and twenty hours (for differentiation) after transfection in NSCs, we performed Western blot for the cell lysates as described previously [13]. Monoclonal mouse GFAP antibody (1:5000), monoclonal mouse MAP2 antibody (1:5000) and monoclonal mouse β-Actin antibody (1:10000) were from CST. The secondary antibodies were goat anti-rabbit (1:10,000) antibody and goat anti-mouse (1:10,000) antibody from CST.
Results
Differential expression of blood exosomal miRNA in MDD
Bioinformatics analysis of the miRNA-seq data resulted in 351 miRNAs for differential analyses between MDD patients and healthy controls (Supplementary Fig. 1A), and showed that 38 blood exosomal miRNAs were DE in MDD patients when compared to controls subjects (Fig. 1a), 24 were upregulated, and 14 were downregulated. To understand the relationship between these DE miRNAs and MDD, we used the well-recognized algorithm microT-CDS [21, 22] to bioinformatically predict the mRNA targets and performed Kyoto Encyclopedia of Genes and Genomes (KEGG) and gene ontology (GO) enrichment analyses to find significantly enriched pathways. The most enriched pathways included axon guidance and development, dendrite, wnt signaling pathway, neuron-to-neuron synapse and PI3K-Akt signaling pathway, which are closely related to depression (Supplementary Fig. 1B, C and Supplementary Table 2).
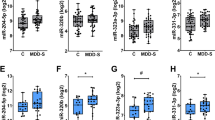
a Heatmap of differentially expressed blood exosomal miRNAs between MDD patients and healthy control subjects. b qRT-PCR validation of has-miR-139-5p expression in the blood exosomes of MDD patients; HC healthy controls, SZ schizophrenia. ROC curves were used to evaluate the accuracy of has-miR-139-5p (c), a cluster of 25 miRNAs (d), and a cluster of ten miRNAs (e) for the diagnosis of MDD; AUC area under the curve. f Dendrogram showing miRNA coexpression modules defined in 79 samples. g Pearson’s correlation coefficient between disease status, age, gender, the Hamilton Depression Rating Scale (HAMD), Montgomery-Asberg Depression Rating Scale (MADRS), and module eigengene. h Log2-transformed fold change distribution of miRNAs in the modules black, blue, brown, magenta, pink, and red.
We validated the top DE miRNA, hsa-miR-139-5p, by qRT-PCR. Our results showed that hsa-miR-139-5p blood exosomal levels were significantly higher in MDD patients than in control subjects, which is consistent with the sequence data (Fig. 1b). The qRT-PCR data showed that the levels of hsa-miR-139-5p did not differ significantly between schizophrenia patients and control subjects, suggesting that the overexpression of this miRNA in blood exosomes is specific to MDD (Fig. 1b).
Blood exosomal miRNAs as biomarkers for MDD diagnosis
We next investigated whether blood exosomal miRNAs could serve as MDD biomarkers. We used the top DE hsa-miR-139-5p to draw the receiver operating characteristic curve and obtained an area under the curve (AUC) of 0.857 (95% CI, 0.769−0.944) for the diagnosis of MDD, with a sensitivity of 0.879 and specificity of 0.717 (Fig. 1c and Supplementary Fig. 2A, B). We then used random forest classifier to find an optimal set of miRNAs that could be used in MDD diagnosis from the 351 miRNAs in blood exosomes, and 25 miRNAs were selected. We used these 25 miRNAs to draw the receiver operating characteristic curve, the AUC to diagnose MDD was 0.966 (95% CI, 0.935–0.998), yielding a sensitivity of 0.970 and specificity of 0.848 (Fig. 1d and Supplementary Fig. 2C, D). We performed an additional curve using the ten miRNAs that contributed most to the differentiation between MDD patients and control subjects, and obtained a sensitivity of 0.939 and specificity of 0.804, the AUC was 0.931 (95% CI, 0.871–0.991) (Fig. 1e and Supplementary Fig. 2e, f).
We then used the sequence data from our previous study [13] to analyze whether these potential blood exosomal biomarkers were MDD-specific. We found that hsa-miR-139-5p had a good performance to differentiate between MDD and schizophrenia patients (Supplementary Fig. 3A–C), and the cluster of above-mentioned 25 miRNAs or 10 miRNAs had an excellent performance to differentiate between MDD and schizophrenia patients (Supplementary Fig. 3D–I), suggesting that these dysregulated blood exosomal miRNAs are MDD-specific.
Perturbation of blood exosomal miRNA coexpression modules in MDD
To further analyze the role of blood exosomal miRNA dysregulation in MDD at a systemic level, we subjected the samples from 33 MDD patients and 46 healthy controls to weighted gene coexpression network analysis (WGCNA) to assign individual miRNAs to coexpression modules and identified ten modules (Fig. 1f). In addition, the analysis showed that disease status was significantly correlated with four modules: two upregulated (blue and red), and two downregulated (magenta and pink) (Fig. 1g, h and Supplementary Fig. 4A−D). We also used WGCNA to assess whether the potential clinical variables (age, gender, and disease severity) were associated with the modules and observed that the MDD-affected modules were not significantly associated with gender, age, HAMD scale, or MADRS scale (Fig. 1g).
We used microT-CDS to predict the four MDD-affected-module targets, and KEGG and GO enrichment analyses were performed to understand the etiology of MDD better. The analyses showed that the top enrichment pathways included axon guidance, axon and dendrite development, glutamatergic synapse, GABAergic synapse, wnt signaling pathway and PI3K-Akt signaling pathway, which suggested a potential role for the MDD-affected modules in disease pathogenesis (Supplementary Fig. 5A–H).
Exosomes from MDD patients caused depressive-like behaviors in mice
To assess the functional involvement of blood exosomes in depression, we isolated blood exosomes from MDD patients and peripherally injected them into normal mice. We found that the mice treated with exosomes from MDD patients showed depressive-like behaviors in the tail suspension (Fig. 2a), forced swimming (Fig. 2b), and novelty suppressed feeding (Fig. 2c) tests, suggesting that exosome is important in the pathogenesis of depression. Exosome treatment did not decrease locomotion in the mice in the open-field test (Fig. 2d), suggesting that the exosome effects were not due to the alteration of locomotion.
Infusion of exosomes isolated from MDD patients into normal mice caused their depressive-like behaviors as assessed by tail suspension test (a), forced swimming test (b), and novelty suppressed feeding test (c). d Traveling distance of different groups of mice in the open-field test. Confocal photomicrographs of DCX-immunostained immature neurons (e) and BrdU/NeuN double-immunostained newborn mature neurons (g) in the dentate gyrus of mice under various treatments. Quantification of DCX-positive cells (f) and newborn mature neurons (h) in the dentate gyrus of mice under various treatments, n = 5. t test. *p < 0.05, **p < 0.01.
We next analyzed oxidative stress marker levels in the mice after exosome treatment. Compared with saline-treated mice, exosome-treated mice had significantly decreased T-AOC activities (p < 0.05) and increased MDA levels (p < 0.05), whereas GSH-PX activity did not show significant difference between saline-treated and exosome-treated mice (Supplementary Fig. 6a–c). We further found that exosome-treated mice had significantly less (p < 0.01) DCX-positive cells in the subgranular zone of the dentate gyrus, a marker of neurogenesis, than saline-treated mice (Fig. 2e, f). Moreover, exosome-treated mice had significantly decreased BrdU/NeuN doubly labeled newborn neurons in the hippocampus when compared with saline-treated mice (Fig. 2g, h).
Antidepressant-like effects of exosomes from healthy subjects in CUMS-treated mice
We next used the CUMS animal model of depression to further assess the functional involvement of blood exosomes in depression. We isolated blood exosomes from healthy human volunteers and peripherally injected them into CUMS-treated mice. We observed that CUMS-treated mice with exosomal treatment showed less depressive-like behaviors in the tail suspension (Fig. 3a), forced swimming (Fig. 3b), and novelty suppressed feeding (Fig. 3c) tests than CUMS-treated mice with saline treatment, suggesting that the exosomes from healthy individuals have antidepressant-like effects. Exosome treatment did not increase locomotion in the open field in the CUMS-treated or control mice (Fig. 3d), suggesting that the exosome effects were not due to the alteration of locomotion. We then analyzed the oxidative stress marker levels in the mice because oxidative stress has been suggested to play a critical role in the pathogenesis of depression [23]. Compared with the control mice, CUMS-treated mice had significantly decreased T-AOC (p < 0.05), GSH-PX (p < 0.05) activities and increased MDA (p < 0.05) levels in the blood (Fig. 3e–g). Exosome treatment significantly increased GSH-PX activity (Fig. 3f, p < 0.05) and decreased MDA levels (Fig. 3g, p < 0.01) in the CUMS-treated mice, whereas T-AOC activity showed a nonsignificant increase in the CUMS-treated mice after the exosome treatment (Fig. 3e, p = 0.074).
CUMS led to depression-like behaviors in mice and administration of blood exosomes form healthy humans reversed the phenotypes as assessed by tail suspension test (a), forced swimming test (b), and novelty suppressed feeding test (c). d Traveling distance of different groups of mice in the open-field test. Effects of exosome treatment on the changes of total antioxidant capacity (T-AOC) (e), glutathione peroxidase (GSH-PX) (f) and malondialdehyde (MDA) (g) in the serum of CUMS mice. h Confocal photomicrographs of DCX-immunostained immature neurons in the subgranular zone of the dentate gyrus of mice under various treatments. i Quantification of DCX-positive cells on the subgranular zone of mice with various treatments, n = 4. j Heatmap of the 274 significantly differentially expressed (DE) proteins in the hippocampus of control and CUMS mice. k Heatmap of the 201 significantly DE proteins in the hippocampus of saline- and exosome-treated CUMS mice. One-way ANOVA with Tukey’s post hoc test. *p < 0.05, **p < 0.01, ***p < 0.001. Data are expressed as mean ± SEM.
Biochemical changes in the hippocampus of CUMS-treated mice with exosome treatment
CUMS-treated mice had significantly less (p < 0.001) DCX-positive cells in the subgranular zone of the dentate gyrus than control mice. Importantly, exosome treatment significantly increased (p < 0.01) DCX-positive cells in CUMS-treated mice (Fig. 3h, i), suggesting that exosome administration promoted adult hippocampal neurogenesis in CUMS-treated mice.
To further understand the molecular pathways underlying the effects of exosome treatment at the central nervous system level, we used proteomics to analyze the hippocampus protein expression profile after various treatments. The LC-MS/MS analysis found 274 DE proteins in the hippocampus of CUMS-treated mice when compared to control mice and 201 DE proteins between saline- and exosome-treated CUMS-treated mice (Fig. 3j, k and Supplementary Fig. 7A). Notably, the expression levels of 38 DE proteins in CUMS-treated mice were restored by exosome treatment (Supplementary Fig. 7A). We then performed KEGG enrichment analyses for the 274 DE and 201 DE proteins, and found nine significantly enriched pathways in both analyses; these included the PI3K-Akt signaling pathway, fatty acid metabolism, and gap junction (Supplementary Fig. 7B, C), therefore supporting the antidepressant-like effects of exosome treatment in mice.
Downregulation of exosomal miR-139-5p alleviates depressive-like behaviors in CUMS mice
We then explored whether exosome-derived miRNA was involved in the antidepressant-like effects of exosomes in the CUMS-treated mice. We observed that CUMS-treated mice had significantly increased blood exosomal miR-139-5p levels when compared with control mice (Fig. 4a), which is consistent with our clinical findings. Importantly, our results demonstrated that brain exosomal miR-139-5p levels, but not whole-brain miR-139-5p levels, were significantly increased in the CUMS-treated mice when compared with control mice (Fig. 4b, c).
a Bar graphs showing the quantification of miR-139-5p levels in the blood exosome of CUMS mice and control mice. b Bar graphs showing the quantification of whole-brain miR-139-5p levels in the CUMS mice and control mice. c Bar graphs showing the quantification of miR-139-5p levels in the brain exosome of CUMS mice and control mice. d Brain exosomal miR-139-5p expression under various treatments in the mice. Intranasal administration of miR-139-5p antagomir reversed depressive-like behaviors in CUMS mice as assessed by tail suspension test (e), forced swimming test (f) and novelty suppressed feeding test (g). h Traveling distance of different groups of mice in the open-field test. Confocal photomicrographs of DCX-immunostained immature neurons (i) and BrdU/NeuN double-immunostained newborn mature neurons (k) in the dentate gyrus of control (I) and CUMS (II) mice treated with vehicle, and control (III) and CUMS (IV) mice treated with miR-139-5p antagomir. Quantification of DCX-positive cells (j) and newborn mature neurons (l) in the dentate gyrus of mice under various treatments, n = 5. a−c t test, d−h, j, l one-way ANOVA with Tukey’s post hoc test. *p < 0.05, **p < 0.01, ***p < 0.001. Data are expressed as mean ± SEM. NC negative control.
To assess the functional involvement of exosomal miR-139-5p upregulation in depressive development of CUMS-treated mice, we intranasally administered 5 nmol miR-139-5p antagomir in the CUMS-treated mice to inhibit exosomal miR-139-5p expression (Fig. 4d). Depressive-like behaviors in the CUMS-treated mice were rescued by intranasal miR-139-5p antagomir administration as assessed by the tail suspension (Fig. 4e), forced swimming (Fig. 4f) and novelty suppressed feeding (Fig. 4g) tests without significant alteration of the locomotive activities (Fig. 4h).
We also found that decreased DCX-positive cells in the dentate gyrus of the CUMS-treated mice was rescued by the intranasal injection of miR-139-5p antagomir (Fig. 4i, j). Furthermore, CUMS-treated mice had significantly decreased BrdU/NeuN doubly labeled newborn neurons in the hippocampus when compared with control mice. However, miR-139-5p antagomir treatment significantly increased newborn neurons in the dentate gyrus of hippocampus (Fig. 4k, l).
MiR-139-5p negatively regulates NSC proliferation
We next directly explored the function of miR-13-5p in cells via in vitro perturbation of miR-139-5p in NSCs (Supplementary Fig. 8A, B). We transfected the NSCs with 100 nM miR-139-5p and analyzed cell proliferation by the WST-1 assay. The results showed that miR-139-5p overexpression inhibited NSC proliferation (Supplementary Fig. 8C). Consistently, silencing miR-139-5p expression by anti-miR-139-5p transfection promoted NSC proliferation (Supplementary Fig. 8D). To validate the effects of miR-139-5p in NSCs, we visually counted neurospheres with altered miR-139-5p expression and found that miR-139-5p overexpression caused a reduction in the size and number of neurospheres, whereas anti-miR-139-5p increased these values (Supplementary Fig. 8E−H).
Since 100 nM miR-139-5p transfection led to a massive overexpression of miR-139-5p in NSCs, we also transfected NSCs with 10 nM miR-139-5p and analyzed cell proliferation by the WST-1 assay (Supplementary Fig. 9A). The results showed that 10 nM miR-139-5p transfection inhibited NSC proliferation (Supplementary Fig. 9B), and caused a reduction in the size and number of neurospheres (Supplementary Fig. 9C, D).
These above results suggest that miR-139-59 is a negative regulator of neural stem cell proliferation.
MiR-139-5p is a negative regulator for neuronal differentiation
To study the role of miR-139-5p in neuronal differentiation in vitro, NSCs were transfected with miR-139-5p and allowed for differentiation. Immunofluorescence data suggested that overexpression of miR-139-5p decreased the number of MAP2-positive cells and increased the number of GFAP-positive cells after NSC differentiation (Fig. 5a). Consistently, anti-miR-139-5p treatment resulted in increased number of MAP2-positive cells and decreased number of GFAP-positive cells after NSC differentiation (Fig. 5b), suggesting that miR-139-5p inhibited the differentiation of NSCs into neurons. In addition, results from Western blot confirmed that miR-139-5p is a negative regulator for neuronal differentiation (Fig. 5c, d).
a Representative confocal pictures (×20, left panel) and quantification (right panel) of MAP2 and GFAP-immunostained cells after miR-139-5p and miR-control treatments in the neural stem cells (NSCs). b Representative ICC confocal pictures (×20) (left panel) and quantification (right panel) of MAP2 and GFAP-immunostained cells after anti-miR-139-5p and anti-miR-control treatments in the NSCs. c Western blot analysis showing downregulation of MAP2 protein expression and upregulation of GFAP protein expression after miR-139-5p treatment in the NSCs. d Western blot analysis showing upregulation of MAP2 protein expression and downregulation of GFAP protein expression after anti-miR-139-5p treatment in the NSCs. e Proposed model illustrating role of exosome-derived miR-139-5p in depressive-like behaviors during stress. a, b n = 15; c, d n = 3. t test, *p < 0.05, **p < 0.01, ***p < 0.001. Data are expressed as mean ± SEM.
Discussion
In this study, we first isolated blood exosomes from MDD patients and provided a comprehensive evaluation of exosomal miRNA dysfunction in MDD. Our miRNA-seq data revealed that MDD patients had a differential expression profile of blood exosomal miRNAs when compared to control subjects. Several previous studies [24, 25] have assessed the changes in blood miRNAs in MDD patients. Yuan et al. [26] systematically reviewed the literature and found that only a few blood miRNAs were consistently dysregulated across studies, the cause of the inconsistency is unknown. Methodological and clinical variables may contribute to heterogeneities between studies. Additionally, given that blood miRNAs are present in apoptotic bodies, exosomes, or as free forms [27], the differential sources of miRNAs in the blood are likely to affect the study results. Although a recent study reported an upregulation of blood levels of whole hsa-miR-361-5p in MDD patients [26], here we provide the evidence showing increased blood levels of exosomal hsa-miR-361-5p in MDD patients. Furthermore, we detected additional 37 DE miRNAs that had not been previously reported, suggesting a relatively unique signature of exosomal miRNA profile in MDD. The hypothesized exosomal miRNA-specific profile in patients with depression was supported by our animal study showing that CUMS-treated mice had elevated miR-139-5p levels in brain exosomes, but not in whole brain.
Different systems in the body, including the immune [28], hematopoietic [29], and nervous systems [30], release exosomes into the blood. However, the percentage of brain-derived exosomes in peripheral blood and whether this percentage is altered in MDD patients is unknown. Here, we propose that at least some dysregulated miRNAs present in the blood exosomes of MDD patients originate during the resorption of cerebrospinal fluid into the venous circulation and that these exosomal miRNAs play critical roles in MDD pathophysiology. This is supported by bioinformatics prediction showing that the targets of MDD-affected miRNAs and miRNA modules were strongly enriched in genes related to synapse, axon and dendrite development, and neurotransmission. Direct evidence of a functional involvement of blood exosomes in the pathogenesis of depression comes from our data showing that transplantation of blood exosomes from MDD patients caused depressive-like behaviors in mice. We then used an animal model of depression and showed that stress-induced depression in mice was rescued after treatment with blood exosomes from healthy human volunteers. It is likely that the disturbed homeostasis in CUMS-treated mice was restored by exosome treatment, and the dysregulated proteins and RNAs in the brains of CUMS-treated mice were at least partially normalized after exosome treatment. This is supported by our proteomics data showing 38 dysregulated proteins in the hippocampus of CUMS-treated mice were restored by exosome treatment. The crucial role of exosomes in depression was further supported by the restoration of hippocampal neurogenesis in CUMS-treated mice after exosome treatment. Additionally, bioinformatics analyses suggested that dysregulated genes were enriched in pathways closely associated with depression, including the PI3K-Akt signaling pathway [31], fatty acid metabolism [32], and gap junction [33]. Therefore, it is likely that the injected blood exosomes crossed the blood−brain barrier to modulate depressive-like behaviors in mice. In fact, Qu et al. [12] showed the ability of blood exosomes to target brain with involvement of the transferrin−transferrin receptor interaction, and intravenous administration of dopamine-loaded exosomes had better therapeutic effect in a Parkinson’s disease mouse model than free dopamine. Additionally, Perets et al. [34] used classical X-ray computed tomography to achieve in vivo neuroimaging of exosomes, and found that intranasally injected exosomes specifically targeted and accumulated in pathologically relevant brain regions of Alzheimer’s disease, Parkinson’ disease and autism animal models. These results suggest that peripheral exosome has the ability to functionally integrate into the brain. However, a limitation of our study is that we cannot exclude the possibility that the injected exosomes targeted molecules present in the peripheral blood that indirectly regulated gene expression in the central nervous system, which then modulated depressive-like behaviors in mice, since exosome treatment diminished the oxidative stress in the peripheral blood of CUMS-treated mice. Nevertheless, our results suggest that exosome is a promising target for the treatment of depression.
The relevance of blood exosome in the pathogenesis of depression was further supported by our data showing that both blood and brain exosomal miR-139-5p levels were significantly upregulated in the CUMS-treated mice, and therefore throwing support for the “peripheral as a window to the brain” hypothesis [35]. Direct evidence of exosome-derived miRNA plays a pivotal role in the onset and/or development of depression is based on our studies using miR-139-5p antagomir. Intranasally administered with miR-139-5p antagomir resulted in the inhibition of heightened expression of brain exosomal miR-139-5p in the CUMS-treated mice, and reduced depression-like behaviors in the stressed mice as measured by forced swimming, tail suspension and novelty suppressed feeding tests, suggesting that exosome-derived miR-139-5p may mediate stress-induced depression. The miR-139-5p antagomir treatment also increased DCX-positive cells (a specific marker of immature neurons) and newborn mature neurons, indicating that miR-139-5p is a negative regulator for neurogenesis. Direct evidence of the regulatory role of miR-139-5p in neurogenesis comes from our in vitro data demonstrating that miR-139-5p is negative regulator for NSC proliferation and neuronal differentiation. The antidepressant-like effects of miR-139-5p antagomir were likely due to the increased neurogenesis in the CUMS-treated mice given that studies have shown that depletion of adult neurogenesis led to depressive-like behaviors in mice [36, 37]. In addition, it has been reported that increased hippocampal neurogenesis was sufficient to mitigate depression-like behaviors in animals [38] and was required for antidepressant efficacy [39]. Therefore, data from these previous studies and together with our present results suggest that stress induces upregulation of exosomal miR-139-5p which results in hippocampal neurogenesis impairment, leading to depressive-like behaviors in mice (Fig. 5e).
Over the last several decades, a large number of studies have aimed to identify biomarkers that could lead to the diagnosis, progression prediction, and treatment of MDD [1, 40]. Although several candidate biomarkers for MDD have been suggested, no relevant biomarker has been validated for clinical use. This is mostly due to sensitivity and specificity issues. For example, these potential biomarkers were later found to be nonspecific for MDD [9]. The data from our study suggested that blood exosomal hsa-miR-139-5p is a promising biomarker for MDD diagnosis since hsa-miR-139-5p showed a good performance in differentiating between MDD patients and healthy controls with a sensitivity of 0.879 and specificity of 0.717. Additionally, our analysis showed that a combination of hsa-miR-139-5p and 24 additional DE blood exosomal miRNAs in MDD improved the diagnostic accuracy with a sensitivity of 0.970 and specificity of 0.848. More importantly, an integrative analysis of our present and previously published data [13] showed that hsa-miR-139-5p and the cluster of 25 miRNAs showed a good/excellent performance in differentiating between MDD and schizophrenia patients, providing further evidence for blood exosomal miRNAs as potential MDD biomarkers.
In conclusion, our genome-wide analysis revealed DE blood exosomal miRNAs in MDD, and indicated that these miRNAs are potential tools for MDD diagnosis. In addition, our study has suggested that exosome played a critical role in the pathogenesis of depression. Based on our data, we proposed that exosome-derived miR-139-5p was upregulated in CUMS-treated mice and resulted in impairment of adult hippocampal neurogenesis, leading to the onset and/or development of depression-like behaviors in mice, which can be rescued by intranasal injection of miR-139-5p antagomir (Fig. 5e). The role of miR-139-5p in neurogenesis was validated by in vitro data showing that miR-139-5p was a negative regulator for NSC proliferation and neuronal differentiation. Therefore, our study suggests a potential new pathway that is activated in the hippocampus during stress to mediate depression-like behaviors in mice, and provides a potential new approach for the diagnosis and treatment of MDD.
Funding and disclosure
This study was supported by the National Natural Science Foundation of China (81703492), Beijing Natural Science Foundation (7182092), High-Level Hospital Development Program for Foshan “Climbing” Project, the Minzu University Research Fund (2018CXTD03), and the MUC 111 project. The authors declare no conflict of interest.
References
Lopez JP, Kos A, Turecki G. Major depression and its treatment: microRNAs as peripheral biomarkers of diagnosis and treatment response. Curr Opin Psychiatry. 2018;31:7–16.
Hamon M, Blier P. Monoamine neurocircuitry in depression and strategies for new treatments. Prog Neuropsychopharmacol Biol Psychiatry. 2013;45:54–63.
Varigonda AL, Jakubovski E, Taylor MJ, Freemantle N, Coughlin C, Bloch MH. Systematic review and meta-analysis: early treatment responses of selective serotonin reuptake inhibitors in pediatric major depressive disorder. J Am Acad Child Adolesc Psychiatry. 2015;54:557–64.
MacQueen G, Santaguida P, Keshavarz H, Jaworska N, Levine M, Beyene J, et al. Systematic review of clinical practice guidelines for failed antidepressant treatment response in major depressive disorder, dysthymia, and subthreshold depression in adults. Can J Psychiatry. 2017;62:11–23.
Hashimoto K. Rapid-acting antidepressant ketamine, its metabolites and other candidates: a historical overview and future perspective. Psychiatry Clin Neurosci. 2019;73:613–27.
Short B, Fong J, Galvez V, Shelker W, Loo CK. Side-effects associated with ketamine use in depression: a systematic review. Lancet Psychiatry. 2018;5:65–78.
Murrough JW, Iosifescu DV, Chang LC, Al Jurdi RK, Green CE, Perez AM, et al. Antidepressant efficacy of ketamine in treatment-resistant major depression: a two-site randomized controlled trial. Am J Psychiatry. 2013;170:1134–42.
Hunsberger JG, Austin DR, Chen G, Manji HK. MicroRNAs in mental health: from biological underpinnings to potential therapies. Neuromolecular Med. 2009;11:173–82.
Fries GR, Quevedo J. Exosomal microRNAs as potential biomarkers in neuropsychiatric disorders. Methods Mol Biol. 2018;1733:79–85.
Steinbichler TB, Dudas J, Riechelmann H, Skvortsova II. The role of exosomes in cancer metastasis. Semin Cancer Biol. 2017;44:170–81.
Tavakolizadeh J, Roshanaei K, Salmaninejad A, Yari R, Nahand JS, Sarkarizi HK, et al. MicroRNAs and exosomes in depression: potential diagnostic biomarkers. J Cell Biochem. 2018;119:3783–97.
Qu M, Lin Q, Huang L, Fu Y, Wang L, He S, et al. Dopamine-loaded blood exosomes targeted to brain for better treatment of Parkinson’s disease. J Control Release. 2018;287:156–66.
Du Y, Yu Y, Hu Y, Li XW, Wei ZX, Pan RY, et al. Genome-wide, integrative analysis implicates exosome-derived microRNA dysregulation in schizophrenia. Schizophr Bull. 2019;45:1257–66.
Vella LJ, Scicluna BJ, Cheng L, Bawden EG, Masters CL, Ang CS, et al. A rigorous method to enrich for exosomes from brain tissue. J Extracell Vesicles. 2017;6:1348885.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Wang L, Li XX, Chen X, Qin XY, Kardami E, Cheng Y, et al. Antidepressant-like effects of low- and high-molecular weight FGF-2 on chronic unpredictable mild stress mice. Front Mol Neurosci. 2018;11:377.
Cheng Y, Rodriguiz RM, Murthy SR, Senatorov V, Thouennon E, Cawley NX, et al. Neurotrophic factor-alpha1 prevents stress-induced depression through enhancement of neurogenesis and is activated by rosiglitazone. Mol Psychiatry. 2015;20:744–54.
Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–62.
MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–8.
Selvaraj P, Xiao L, Lee C, Murthy SR, Cawley NX, Lane M, et al. Neurotrophic factor-alpha1: a key Wnt-beta-Catenin dependent anti-proliferation factor and ERK-Sox9 activated inducer of embryonic neural stem cell differentiation to astrocytes in neurodevelopment. Stem Cells. 2017;35:557–71.
Coutinho de Almeida R, Ramos YFM, Mahfouz A, den Hollander W, Lakenberg N, Houtman E, et al. RNA sequencing data integration reveals an miRNA interactome of osteoarthritis cartilage. Ann Rheum Dis. 2019;78:270–7.
Wu C, Zhao Y, Liu Y, Yang X, Yan M, Min Y, et al. Identifying miRNA-mRNA regulation network of major depressive disorder in ovarian cancer patients. Oncol Lett. 2018;16:5375–82.
Jimenez-Fernandez S, Gurpegui M, Diaz-Atienza F, Perez-Costillas L, Gerstenberg M, Correll CU, et al. Oxidative stress and antioxidant parameters in patients with major depressive disorder compared to healthy controls before and after antidepressant treatment: results from a meta-analysis. J Clin Psychiatry. 2015;76:1658–67.
Maffioletti E, Cattaneo A, Rosso G, Maina G, Maj C, Gennarelli M, et al. Peripheral whole blood microRNA alterations in major depression and bipolar disorder. J Affect Disord. 2016;200:250–8.
Roy B, Dunbar M, Shelton RC, Dwivedi Y. Identification of microRNA-124-3p as a putative epigenetic signature of major depressive disorder. Neuropsychopharmacology. 2017;42:864–75.
Yuan H, Mischoulon D, Fava M, Otto MW. Circulating microRNAs as biomarkers for depression: many candidates, few finalists. J Affect Disord. 2018;233:68–78.
Zhang J, Li S, Li L, Li M, Guo C, Yao J, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteom Bioinforma. 2015;13:17–24.
Tan L, Wu H, Liu Y, Zhao M, Li D, Lu Q. Recent advances of exosomes in immune modulation and autoimmune diseases. Autoimmunity. 2016;49:357–65.
Liao FL, Tan L, Liu H, Wang JJ, Ma XT, Zhao B, et al. Hematopoietic stem cell-derived exosomes promote hematopoietic differentiation of mouse embryonic stem cells in vitro via inhibiting the miR126/Notch1 pathway. Acta Pharm Sin. 2018;39:552–60.
Budnik V, Ruiz-Canada C, Wendler F. Extracellular vesicles round off communication in the nervous system. Nat Rev Neurosci. 2016;17:160–72.
Marsden WN. Synaptic plasticity in depression: molecular, cellular and functional correlates. Prog Neuropsychopharmacol Biol Psychiatry. 2013;43:168–84.
Fernandes MF, Mutch DM, Leri F. The relationship between fatty acids and different depression-related brain regions, and their potential role as biomarkers of response to antidepressants. Nutrients. 2017;9:298.
Ren Q, Wang ZZ, Chu SF, Xia CY, Chen NH. Gap junction channels as potential targets for the treatment of major depressive disorder. Psychopharmacology. 2018;235:1–12.
Perets N, Betzer O, Shapira R, Brenstein S, Angel A, Sadan T, et al. Golden exosomes selectively target brain pathologies in neurodegenerative and neurodevelopmental disorders. Nano Lett. 2019;19:3422–31.
Qin XY, Cao C, Cawley NX, Liu TT, Yuan J, Loh YP, et al. Decreased peripheral brain-derived neurotrophic factor levels in Alzheimer’s disease: a meta-analysis study (N = 7277). Mol Psychiatry. 2017;22:312–20.
Egeland M, Guinaudie C, Du Preez A, Musaelyan K, Zunszain PA, Fernandes C, et al. Depletion of adult neurogenesis using the chemotherapy drug temozolomide in mice induces behavioural and biological changes relevant to depression. Transl Psychiatry. 2017;7:e1101.
Snyder JS, Soumier A, Brewer M, Pickel J, Cameron HA. Adult hippocampal neurogenesis buffers stress responses and depressive behaviour. Nature. 2011;476:458–61.
Hill AS, Sahay A, Hen R. Increasing adult hippocampal neurogenesis is sufficient to reduce anxiety and depression-like behaviors. Neuropsychopharmacology. 2015;40:2368–78.
Eisch AJ, Petrik D. Depression and hippocampal neurogenesis: a road to remission? Science. 2012;338:72–75.
Gadad BS, Jha MK, Czysz A, Furman JL, Mayes TL, Emslie MP, et al. Peripheral biomarkers of major depression and antidepressant treatment response: current knowledge and future outlooks. J Affect Disord. 2018;233:3–14.
Acknowledgements
We thank Dr. X. Zhang for critical comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Wei, ZX., Xie, GJ., Mao, X. et al. Exosomes from patients with major depression cause depressive-like behaviors in mice with involvement of miR-139-5p-regulated neurogenesis. Neuropsychopharmacol. 45, 1050–1058 (2020). https://doi.org/10.1038/s41386-020-0622-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41386-020-0622-2
This article is cited by
-
Exosome-mediated repair of spinal cord injury: a promising therapeutic strategy
Stem Cell Research & Therapy (2024)
-
Prevalence and risk factors for subclinical hypothyroidism in older patients with major depressive disorder
BMC Geriatrics (2024)
-
Exosomes: potential targets for the diagnosis and treatment of neuropsychiatric disorders
Journal of Translational Medicine (2024)
-
Current progression in application of extracellular vesicles in central nervous system diseases
European Journal of Medical Research (2024)
-
Multiomics and blood-based biomarkers of electroconvulsive therapy in severe and treatment-resistant depression: study protocol of the DetECT study
European Archives of Psychiatry and Clinical Neuroscience (2024)