Abstract
Standardized programmed death-ligand 1 (PD-L1) assessment in non-small cell lung cancer (NSCLC) is challenging, owing to inter-observer variability among pathologists and the use of different antibodies. There is a strong demand for the development of an artificial intelligence (AI) system to obtain high-precision scores of PD-L1 expression in clinical diagnostic scenarios. We developed an AI system using whole slide images (WSIs) of the 22c3 assay to automatically assess the tumor proportion score (TPS) of PD-L1 expression based on a deep learning (DL) model of tumor detection. Tests were performed to show the diagnostic ability of the AI system in the 22c3 assay to assist pathologists and the reliability of the application in the SP263 assay. A robust high-performance DL model for automated tumor detection was devised with an accuracy and specificity of 0.9326 and 0.9641, respectively, and a concrete TPS value was obtained after tumor cell segmentation. The TPS comparison test in the 22c3 assay showed strong consistency between the TPS calculated with the AI system and trained pathologists (R = 0.9429–0.9458). AI-assisted diagnosis test confirmed that the repeatability and efficiency of untrained pathologists could be improved using the AI system. The Ventana PD-L1 (SP263) assay showed high consistency in TPS calculations between the AI system and pathologists (R = 0.9787). In conclusion, a high-precision AI system is proposed for the automated TPS assessment of PD-L1 expression in the 22c3 and SP263 assays in NSCLC. Our study also indicates the benefits of using an AI-assisted system to improve diagnostic repeatability and efficiency for pathologists.
Similar content being viewed by others
Introduction
Companion diagnostic immunohistochemistry (IHC) tests assessing programmed death-ligand 1 (PD-L1) expression are widely used to predict the response to immunotherapy as a biomarker1,2,3. PD-L1 scoring the percentage of PD-L1-positive tumor cells (%TC) is commonly recommended for immunotherapeutic categories in several antibodies used in non-small cell lung cancer (NSCLC)4. Immunotherapy development has increased the requirements for the precise assessment of immune biomarkers that enable patient selection5. However, the interpretation of PD-L1 IHC staining is a difficult task because of the continuously variable nature of PD-L1 expression, which is characterized by intratumoral heterogeneity, positivity on various cell types, and various antibodies with different possible staining properties6,7,8. Inter-observer variability and a lack of reproducibility between observations are some of the common challenges in clinical practice and may contribute to inappropriate patient stratification9,10,11. In addition, the calculation of PD-L1 expression is a time-consuming process that increases the routine workload of pathologists.
Advances in digital pathology and widely available slide scanners have set the foundation for the clinical application of artificial intelligence (AI) to develop assisted computer tools for improving pathologic practice12,13,14. Image-processing AI has been developed by expert systems to machine learning (ML) and deep learning (DL) in recent decades15. DL has been used for several whole slide image (WSI) analysis tasks, including histopathological diagnosis16,17, cancer detection18, cell classification and enumeration19, genomic mutation and microsatellite instability prediction20,21, tumor grade22, and cancer prognostication23. The use of digital approaches improves the quantitative diagnosis of IHC staining, such as that of human epidermal growth factor receptor 2, indicating the potential of AI in assisting pathologists with tumor-specific biomarkers for stratified medicine24,25,26.
Several studies have used the WSI of PD-L1 slides and manual supervision to show that image-based scoring algorithms are highly consistent with pathologists27,28,29. However, limited strategies for automated PD-L1 scoring have been proposed using deep learning algorithms and computer approaches. Developing clinically acceptable AI systems to automatically obtain high-precision scores on PD-L1 antibodies and assist pathologists in clinical settings remains challenging.
In this study, we proposed a DL-based computational system to automatically detect the tumor area of NSCLC and to calculate the tumor proportion score (TPS) of PD-L1 in FDA-approved 22c3 and SP263 assays. The TPS assessed using the AI system was compared with that computed by the pathologists. The aim of this study was to develop an AI system approach to assist pathologists in the precise diagnosis of PD-L1 expression in NSCLC.
Material and methods
Datasets
The datasets comprised the following five parts: (1) The DL model training set contained 173 WSIs of the 22c3 assay for training the DL model for tumor area detection, including 133 WSIs of resected lung adenocarcinoma (LADC) and the remaining resected lung squamous cell carcinomas (LSQCs), and approximately 1–5 representative tumor areas (>1 mm2) were selected from each WSI of the 22c3 assays for further model training. To obtain a robust model, the tumor regions were evaluated by specialist pathologists and selected with a balance of subtypes, especially in LADC, which comprises various histological subtypes. One or two representative non-neoplastic regions (>1 mm2), including various immune cells (macrophages and lymphocytes), hemorrhage, necrosis, and stromal cells, were also selected as negative inputs. A total of 563 regions were selected for the DL model training set. (2) The DL model test set contained 114 selected regions of 78 WSIs of the 22c3 assay and was used to evaluate the performance of tumor area prediction and to determine the most appropriate structure of the DL model. (3) The TPS comparison test set contained 110 slides (55 resected samples and 55 biopsy samples) and was used to test the consistency of TPS between that obtained by the AI system and pathologists in the 22c3 assay; 111 regions were selected from 61 WSIs to test the tumor detection performance. (4) An AI system-assisted test set contained 40 WSIs, including 20 resected samples and 20 biopsy samples, and was used to test the performance of the AI-assisted procedure of the TPS analysis for pathologists in the 22c3 assay. (5) The Ventana PD-L1 (SP263) assay test set contained 100 WSIs, including 50 resected samples and 50 biopsy samples, and was used to test the performance of the AI system in tumor detection and TPS evaluation in the SP263 assay; 114 regions were selected from 61 WSIs.
All 22c3 IHC slides were selected from a collection of clinical detection specimens at Peking University Cancer Hospital (PKUCH) and Tianjin Medical University Cancer Hospital (TMUCH) from January 2018 to December 2019, whereas SP263 IHC slides were collected from a collection of experimental studies at PKUCH from October 2019 to December 2019. The detailed dataset information is summarized in Tables S1 and S2.
PD-L1 staining and TPS interpretation by pathologists
PD-L1 staining and WSI preparation
All slides were obtained by cutting formalin-fixed, paraffin-embedded NSCLC samples into 4 μm-thick sections and stained with PD-L1 IHC 22c3 pharmDx assay (Dako, Carpenteria, CA, USA) according to the standard staining protocol using the Dako Autostainer Link 48 platform. PD-L1 IHC SP263 assays (Ventana Medical Systems, Tucson, AZ, USA) were developed on the Ventana BenchMark platform. All PD-L1 IHC slides were scanned using a Leica Aperio CS2 (Leica Biosystems, Buffalo Grove, IL, USA) at 20× magnification.
TPS interpretation by pathologists
TPS was estimated as the percentage of viable TCs showing partial or complete membrane staining5. To obtain more detailed comparative data, the TPS values calculated by pathologists were recorded as follows: <1, 1–2, 3–5, 5–9, 10–14, 15–19, … 90–94, 95–98, and 99–100%, with intervals of 5% between 5 and 95% for each range. As described in the protocols, TPS of <1% was considered negative, 1–49% was considered low expression, and ≥50% was considered high expression. In this study, “TPS-pathologists” refers to TPS estimated by pathologists, “TPS-trained” to TPS estimated by trained pathologists, and “TPS-untrained” to TPS estimated by untrained pathologists.
AI system development
Annotation procedure
All regions (tumor and stroma) in the datasets were annotated by five pathologists using QuPath (Version 0.2.2)30. A pilot study was performed to test the concordance of the labels from different pathologists. The intersection over union (IoU) and concordance among pathologists on three random slides were tested (Table S3). The regions were then randomly assigned to five pathologists after comparing the concordance of the manual annotations. To obtain high-quality datasets, the labels were reviewed by a consultant pathologist to resolve any controversy in the annotation procedure.
DL model development
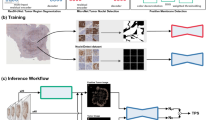
Figure 1a illustrates the process of the entire AI system. A DL model was built based on a U-Net structure, with residual blocks in the model to accelerate convergence and avoid degradation of the deep neural network. U-Net, which displays excellent performance in the field of image segmentation, was employed in PD-L1 IHC WSIs for tumor area detection. Our DL model contains an encoding component to extract features and a follow-up decoding component to restore the image and classify each pixel (Fig. 1b). In the training procedure, the DL model was trained to simultaneously segment the tumor area and classify the input region as auxiliary loss. Only the output of the tumor area segmentation task was used to predict the tumor area.
a The design of this study was to develop an artificial intelligence (AI) system to automatically assess the tumor proportion score (TPS) of PD-L1 expression in non-small cell lung cancer (NSCLC). b DL model development consisting of an annotation dataset and U-Net training for testing tumor detection performance. c Histograms of DL model performance in NSCLC and subtypes of lung adenocarcinoma (LADC) and lung squamous cell carcinoma (LSQC) in the DL model test set (22c3 assay).
WSIs with a resolution of 20× magnification were used for training and testing the DL model. 256 × 256 pixel patches were randomly obtained from the WSI annotation areas and fed into the network for model training. The categorization labels during classification model training were set according to whether the proportion of the tumor area in each patch was greater than 0.75. To obtain a high-performance model, data enhancements were performed during the training. Because the PD-L1 IHC slides were collected from different hospitals and may have different staining properties, the color of the images was also enhanced. After processing by a trained deep neural network model, each pixel in the image was assigned a probability value to determine whether it was in the tumor area (>0.5) or not. In the prediction process, a sliding window (256 × 256 pixels, step size of 128 pixels) was used; however, only the prediction results in the middle of the window (128 × 128 pixels) were recorded.
Cell segmentation and TPS calculation
For tumor area detection using the DL model, the tumor cells were segmented with a modified watershed algorithm, which is a common method for peak detection in cell segmentation, in QuPath (Version 0.2.2)30. The TPS calculation process requires a cell-segmentation threshold and a positive discrimination threshold. Before being applied to the entire dataset, parameter initialization was performed on a randomly chosen group of 3–5 WSIs. We searched for the best default thresholds for the dataset with this group based on the specialist pathologists’ observations of the visualized cell segmentation and positive discrimination results. The entire dataset was processed with the adjusted thresholds, and the TPS was calculated using the number of PD-L1-positive and -negative cells, as the ratio of the number of PD-L1-positive tumor cells to the total number of tumor cells. In this study, “TPS-AI” was defined as the TPS calculated by the proposed AI system.
AI system visualization
Each IHC image showed the tumor detection results and enabled pathologists to visualize the reliability of TPS evaluation by the AI system. Additionally, the segmented cells were marked on IHC images with different colors representing PD-L1-positive or -negative status to show positive discrimination results. In the current AI system, PD-L1-positive and -negative tumor cells are shown as red and green dots, respectively. Thus, PD-L1 pathologists could easily identify PD-L1-positive or -negative tumor cells to confirm the TPS calculation.
TPS comparison test in the 22c3 assay
Six pathologists, including three highly trained pathologists (PD-L1 22c3 assay certified) and three untrained pathologists, were involved in this study using the TPS comparison test set. The TPS assessed by the AI system was compared with the median and average results of TPS evaluated under a microscope by pathologists. The test time was recorded. The TPS results from three trained pathologists were used as the ground truth. The consistency of TPS in LADC and LSQC subtypes, specimen types, and the two institutes was analyzed.
AI system-assisted diagnosis test in the 22c3 assay
In the AI-assisted diagnosis test, six pathologists evaluated the TPS of PD-L1 22c3 IHC expression using the AI system-assisted test set. Three trained pathologists evaluated the TPS only with WSIs on a computer, and three untrained pathologists evaluated the TPS with WSIs, which contained the pre-read results and visualized figures by the proposed AI system to mimic a computer-aided diagnosis workflow. The time cost was recorded for each participant with and without the aid of the AI system.
Ventana PD-L1 (SP263) assays test
The DL model, which was trained on the 22c3 assay, was directly tested on SP263 assays for its performance in tumor detection without additional training or transfer learning with the Ventana PD-L1 (SP263) assay test set. TPS was automatically calculated on SP263 WSIs using the same procedure as the 22c3 WSIs. Two trained pathologists (J. W. and X. Y.) independently evaluated PD-L1 (SP263) IHC slides under a microscope. The inconsistent cases were reviewed by a third consultant pathologist (D. L.) to obtain a consensus. The TPS of the SP263 assay in this test, which was assessed by the AI system, was compared with the consensus results obtained by the pathologists.
Evaluation metrics
Metrics (including precision, recall, specificity, F1-score, IoU, and accuracy) were used in this study to evaluate the performance of the DL model for tumor detection.
Statistics
The intraclass correlation coefficient (ICC) was used when TPS was treated as a continuous variable among the pathologists. Pearson’s correlation coefficient was used for comparison with TPS-AI and the median and average continuous variables of the TPS-pathologists. Fleiss’ Kappa was calculated for inter-pathologist agreement, and Cohen’s kappa was calculated for the agreement of the interval variables of TPS-pathologists and TPS-AI. The Kappa values were categorized as poor (<0.40), moderate (0.40–0.70), or excellent (≥0.70). Statistical significance was set at p < 0.05. All statistical analyses were performed using Python (version 3.6) with scikit-learn and scipy.
Results
Model training and performance in the 22c3 assay
The DL model was iteratively trained and subsequently evaluated on the test set to demonstrate its ability to detect tumor areas. The DL model achieved high performance on PD-L1 IHC WSIs. For all cases of NSCLC, the results showed an accuracy of 0.9326, specificity of 0.9641, precision of 0.9248, recall of 0.8609, F1-score of 0.8871, and an IoU of 0.8051. The DL model also showed high performance in the test sets of the LADC and LSQC subtypes. LADCs have more complex histological structures than LSQCs and are usually divided into several predominant subtypes. Additionally, the performance of the DL model was evaluated on these subtypes, and the model showed similarly high accuracy and specificity in LADC subtypes. In particular, high specificity was obtained in both LADC (0.9681) and LSQC (0.9549) and maintained over 0.95 in any subtype of LADC to minimize the false detection of non-neoplastic cells that possibly showed PD-L1 co-expression and influenced the TPS output. Therefore, a highly specific and highly accurate DL model that enables automated detection of the tumor area was prepared for the calculation of TPS on PD-L1 WSIs. Figure 1c and Table S4 show the performance of the experimental DL model.
TPS comparison test in the 22c3 assay
Based on tumor area detection of PD-L1 IHC WSIs by the DL model, TPS was automatically calculated with positivity analysis after tumor cell segmentation and DAB baseline evaluation. The TPS comparison test set was composed of 110 WSIs, including 55 surgical samples and biopsies from each NSCLC. Figure 2, S1, and S2 show examples of the visualization of tumor area detection and TPS calculation in PD-L1 IHC WSIs for the surgical samples and biopsies, respectively.
Visualization of typical examples of PD-L1 scoring in non-small cell lung cancer (NSCLC) From left to right: PD-L1 immunohistochemistry (IHC) raw images, tumor detection, and PD-L1 calculation. PD-L1-positive and -negative tumor cells were set as red and green dots in the PD-L1 calculation, respectively.
In this test set, the DL model showed high performance in tumor area detection with an accuracy of 0.9464, specificity of 0.9594, precision of 0.9318, recall of 0.8882, F1-score of 0.9063, and an IoU of 0.8344 (Fig. 3a and Table S4).
a Histograms of deep learning (DL) model performance in non-small cell lung cancer (NSCLC) and subtypes of lung adenocarcinoma (LADC) and lung squamous cell carcinoma (LSQC) in the tumor proportion score (TPS) comparison test set. b The intraclass consistency of TPS in trained and untrained pathologists. c Consistency of TPS between AI system (TPS-AI) and average and median TPS in trained pathologists (TPS-trained) and untrained pathologists (TPS-untrained). d TPS agreement among pathologists and AI system using intervals applied in current clinical practice.
To test the performance of the AI system in the TPS calculation, six pathologists, including three trained pathologists and three untrained pathologists, examined the slides using a microscope. Initially, the intraclass consistency of TPS-pathologists in subgroups of trained and untrained pathologists was compared. The trained and untrained pathologists obtained intraclass consistencies with ICC values of 0.963 (95% confidence interval [CI], 0.941–0.977) and 0.922 (95% CI, 0.879–0.952), respectively) (Fig. 3b, Table 1, and Table S5). During the test, the untrained pathologists had a longer average time per slide than the trained pathologists (2.58 vs. 2.15 min).
The TPS-AI and TPS-pathologists were then compared to show the consistency between the AI system and pathologists; the average and median variables of TPS-pathologists were used in the comparison test. The results showed a high consistency between the TPS-AI and TPS-trained (TPS-AI vs. average TPS-trained: R = 0.9458; TPS-AI vs. median TPS-trained: R = 0.9429). The consistency between TPS-AI and TPS-untrained was analyzed and found to be relatively lower (TPS-AI vs. average TPS-untrained: R = 0.9087; TPS-AI vs. median TPS-untrained: R = 0.8878) than that of trained pathologists (Fig. 3c and Table 1).
Surgical samples and biopsies are commonly used in clinical processes for PD-L1 detection and are included in the comparison test. The TPS-AI in biopsies was highly consistent with that of trained pathologists (TPS-AI vs. average TPS-trained: R = 0.9794; TPS-AI vs. median TPS-trained: R = 0.9763), whereas a relatively lower consistency was obtained in surgical samples (TPS-AI vs. average TPS-trained: R = 0.8990; TPS-AI vs. median TPS-trained: R = 0.8968). The performance of the AI system in PD-L1 IHC WSIs from the two centers was also considered for possible staining differences. The AI system achieved a similarly high performance in samples from both centers: for PKUCH, TPS-AI vs. average TPS-trained: R = 0.9444; TPS-AI vs. median TPS-trained: R = 0.9407; for TMUCH, TPS-AI vs. average TPS-trained: R = 0.9440; TPS-AI vs. median TPS-trained: R = 0.9477. High consistency for the NSCLC subtypes was achieved in both LADC and LSQC: for LADC, TPS-AI vs. average TPS-trained: R = 0.9373; TPS-AI vs. median TPS-trained: R = 0.9333; for LSQC, TPS-AI vs. average TPS-trained: R = 0.9680; TPS-AI vs. median TPS-trained: R = 0.9658 (Fig. S3 and S4 and Tables S6).
In the current clinical PD-1/PD-L1 immunotherapy procedure, TPS intervals of the 22c3 assay are commonly recommended at a cutoff of 1 and 50% as thresholds for patient stratification for potential pembrolizumab benefits. Therefore, the TPS-AI and TPS-pathologists were compared using cutoffs at 1 and 50%. The results showed that the intraclass agreement of untrained pathologists were also lower than those of trained pathologists. When comparing the TPS-AI and TPS-pathologists, moderate and excellent agreement was obtained at kappa values of approximately 0.6 at TPS < 50% and 0.898 at TPS ≥ 50%, respectively. Figure 3c, S3, and S4 and Tables S7 and S8 show the results of the comparison of TPS-AI and TPS-pathologists in the categorized TPS.
AI system-assisted diagnosis test
The TPS comparison test showed that the proposed AI system was highly consistent with trained pathologists in the 22c3 assay. The AI-assisted diagnosis tests (schematic shown in Fig. S5) were then performed to reveal the potential impact on pathologists when evaluating TPS with the assistance of an AI system with a pre-diagnostic analysis and visualization of PD-L1 positive/negative tumor cells. The intraclass consistency among pathologists was compared; the ICC of untrained pathologists who assisted with the AI system was higher than that of trained pathologists who calculated the TPS only with WSIs on a computer without AI assistance (0.944 in TPS-untrained vs. 0.866 in TPS-trained) and higher than the diagnostic procedure of untrained pathologists under the microscope (ICC of TPS-untrained, 0.922) (Fig. 4a and Table S9). Comparison of the TPS results between pathologists and AI system showed a higher consistency between the untrained pathologists and AI system in this test (average of 0.9799 for TPS-untrained vs. an average of 0.8506 for TPS-trained; 0.9701 in median TPS-untrained vs. 0.8551 in median TPS-trained) (Table 1). Similarly, higher intraclass agreement and higher accordance between TPS-AI and TPS-pathologists were also achieved in untrained pathologists in the categorized TPS (Fig. S6 and Table S7).
a The intraclass consistency among trained pathologists diagnosing whole slide images (WSIs) and untrained pathologists diagnosing WSIs assisted by the AI system. b The AI system significantly reduces the time cost and improves the intraclass consistency for tumor proportion score (TPS) evaluation by untrained pathologists. c Histograms of deep learning (DL) model performance in non-small cell lung cancer (NSCLC) and subtypes of lung adenocarcinoma (LADC) and lung squamous cell carcinoma (LSQC) in the Ventana PD-L1 (SP263) assay test set. d Consistency between TPS-AI and TPS-pathologists in the SP263 assay. e TPS agreement among pathologists and the AI system using intervals in the SP263 assay.
In this test, all six pathologists evaluated the TPS using WSIs; the three untrained pathologists spent less time than the trained pathologists. The three trained pathologists took an average time of 2.85 min per WSI (ranging from 2.70 to 3.03 min), whereas the three untrained pathologists took 1.38 min per WSI (ranging from 1.30–1.48 min). With the assistance of the AI system, the time taken for untrained pathologists was less than that of trained pathologists, and less than the diagnosis process of PD-L1 evaluation by trained/untrained pathologists under the microscope (Fig. 4b and Table S10). Therefore, the test indicated that the AI system improved the repeatability and significantly reduced the time cost of TPS evaluation for untrained pathologists.
Model performance and TPS comparison on Ventana PD-L1 (SP263) assays
The DL model was only trained using the 22c3 assay; therefore, the AI system was also tested for its performance in Ventana PD-L1 (SP263) assays to explore the reliability of a different antibody. A similar performance of DL models in the SP263 dataset was achieved (accuracy: 0.9624; specificity: 0.9793; precision: 0.9265; recall: 0.8848, F1-score: 0.9025; and IoU: 0.8307) (Fig. 4c and Table S4). High specificity and accuracy were achieved for both LADC and LSQC (Table S4). Thus, the DL model was robust for tumor area detection using SP263 assays.
TPS was further calculated using the AI system on 100 WSIs of SP263 IHC staining and compared with the TPS results of consensus from specialist pathologists. The results showed a high consistency between the TPS-AI and TPS-pathologists (TPS-AI vs. TPS-pathologists: R = 0.9787) (Fig. 4d and Table 1). Similarly, high consistency was obtained in surgical samples and biopsies and in LADC and LSQC (Fig. S7 and Table S6). In addition, the TPS-AI and TPS-pathologists were compared using cutoffs at 1 and 50%. The results showed moderate and excellent agreement of approximately 0.65 at TPS < 50% and 0.926 at TPS ≥ 50%, respectively (Fig. 4e and Table S7). Figure S8 shows examples of the visualization of the tumor area detection and TPS calculation in SP263.
Discussion
We developed a fully automated method that enables tumor area detection and quantitative scoring of TPS in digital PD-L1 IHC slides of NSCLC. This system displayed high consistency in the TPS evaluation of 22c3 (R = 0.9429–0.9458) and SP263 (R = 0.9787) assays between the AI system and trained pathologists. Moreover, the potential benefits of the AI system in improving the efficiency and interobserver repeatability of pathologists were demonstrated.
Precise assessment of PD-L1 IHC expression requires specialized training, considerable expertise, and a longer practice time for pathologists. Consistent with the findings of previous studies, highly trained pathologists showed higher intraclass consistency compared to untrained pathologists31. TPS from highly trained pathologists with high intraclass consistency as a measurement of the ground truth was utilized. The AI system obtained a higher consistency with trained pathologists than untrained pathologists (R = 0.963 vs. 0.922). Therefore, the diagnostic ability of the system could be as capable as trained pathologists and possibly higher than that of general pathologists. Koelzer et al. trained a PD-L1 scoring algorithm for melanomas using random forest after manual annotation and showed a high concordance with the %TC scores generated by pathologists (R = 0.97)27. We achieved a performance similar to that of a fully automated method that did not require manual annotation to achieve successful PD-L1 analysis.
As TPS counting was based on PD-L1 membrane positivity on TCs, distinguishing PD-L1 positive on non-neoplastic cells can be challenging in PD-L1 %TC scoring. Immune cells (e.g., macrophages and lymphocytes) also commonly express PD-L1, which may exert an influence on %TC scoring when not clearly excluded in cancer detection. We used supervised learning comprising U-Net and residual blocks and trained the model with a pilot study to standardize the tumor labels from different pathologists. To minimize the potential influence of the non-neoplastic cells that could co-express PD-L1, we annotated multiple non-neoplastic areas as negative inputs and developed a highly specific DL model for TC detection (>0.95 in all NSCLC subtypes) for use in the calculation of PD-L1 on TCs. High-performance tumor area detection and cell segmentation models are crucial for accurate TPS interpretation.
In the actual use scenario, the AI system was designed to automatically process tumor detection, cell segmentation, and positive discrimination after the pathologists uploaded WSIs into the system. The system assists pathologists by pre-reading PD-L1 slides and presenting quantitative values as second opinions to pathologists. Furthermore, visualization of the AI assistance system is useful for pathologists during PD-L1 diagnosis. The AI system marks the tumor area after detection and PD-L1-positive and -negative tumor cells after segmentation on the images. The images with marks would be provided for pathologists to assist them in rapidly performing a scrutinized assessment and reducing the pathology workload by automatically highlighting PD-L1-positive TCs.
General pathologists commonly showed higher inter-observer variability and took a longer time than highly trained pathologists in a conventional TPS interpretation process31. We performed an AI system-assisted diagnosis test to reveal the impact of AI approaches on general pathologists. By comparing the intraclass consistency and time cost between the untrained pathologists who assisted with the AI system and the trained pathologists without AI assistance, we observed a higher ICC and a lower time cost in the subgroup of untrained pathologists. This test demonstrated the potential benefits of the AI system to help pathologists improve the repeatability and efficiency by providing pre-read PD-L1 results and the visualization of PD-L1 expression cells.
In current clinical practice, several assays for detecting PD-L1 expression using IHC analysis have been developed for different platforms32. Pathologists need standardized training for different antibodies, which significantly increases the complexity of PD-L1 interpretation in practice33. We first demonstrated the generalization of the AI tool using different PD-L1 antibodies. To explore the reliability of the AI system with different antibodies, direct testing of the DL model on SP263 without additional transfer learning was performed; a similarly high performance was achieved in the tumor area. Based on the characteristics of SP263 staining, the DAB default baseline of SP263 was reset, and the TPS assessed by the AI system was consistent with that of specialist pathologists, indicating that the AI system can also be used for SP263 assay analysis. This DL model showed potential generalization to different antibodies, which may aid in the acceleration of the standardization of different antibody protocols. Harmonization of PD-L1 evaluation in different assays may help standardize patient screening and accelerate clinical trials and the application of PD1/PD-L1 target immunotherapy6.
The inconsistency between the AI system and pathologists was mainly due to occasional poor IHC staining and different judgments of scattered tumor cells (Fig. S9). Overstaining, cytoplasmic positivity, or weak staining were the most common causes of abnormal PD-L1 expression analysis. The scattered macrophages that intermix in tumor nests were not easily identified. Such cases are commonly controversial among pathologists, and the consistency between pathologists and the AI system is also relatively lower at a cutoff at 1%. The cell segmentation and DAB parameter threshold must be actively adjusted by pathologists in such situations. In this study, AI system testing in the SP263 assay obtained a slightly higher performance than the 22c3 assay for the SP263 dataset collected from an experimental study with higher staining quality and homogeneity than the 22c3 dataset, which was collected from routine clinical detection. Weak staining of PD-L1, which could be highlighted by the AI system, might not be observed by pathologists during diagnosis. Thus, the AI system could reduce the omission of weakly positive expression for pathologists in routine diagnosis.
This study had some limitations. First, only two institutes were involved, and the dataset was relatively small. More datasets collected from multiple institutions would increase the robustness of the AI model, and a multicenter clinical trial is required to verify the performance of the AI system in real-world settings. Second, the DL-based tumor detection model, which occasionally misidentifies non-neoplastic cells, may cause a certain deviation in TPS outputs. Third, the AI system was tested only on approved 22c3 and SP263 assays. The system will be validated on more antibodies when adequate slides from other assays are available. Finally, because of complex clinical situations, none of the DL models always detect all rare subtypes of NSCLC, which may lead to misjudgment of the tumor area in rare cases. Visualization of quality control is necessary for the clinical application of AI tools, and a Man-Machine integration34 in the PD-L1 diagnostic workflow is recommended.
In conclusion, our study extended tumor area detection, automated PD-L1 scoring %TCs, and generalization of different antibodies. The AI system achieved high consistency with specialist pathologists in the TPS analysis of both the 22c3 and SP263 assays. The diagnostic repeatability and efficiency could be improved with the pre-reading quantitative results and visualization of PD-L1 expression status provided by the AI system. This study indicated that the AI-assisted system could be an effective and valuable tool to overcome the challenges of PD-L1 assessment in the field of immunotherapy.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Garon, E. B. et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med. 372, 2018–2028 (2015).
Reck, M. et al. Updated analysis of KEYNOTE-024: pembrolizumab versus platinum-based chemotherapy for advanced non-small-cell lung cancer with PD-L1 tumor proportion score of 50% or greater. J. Clin. Oncol. 37, 537–546 (2019).
Yu, H., Boyle, T. A., Zhou, C., Rimm, D. L. & Hirsch, F. R. PD-L1 expression in lung cancer. J. Thorac. Oncol. 11, 964–975 (2016).
Blank, C. U., Haanen, J. B., Ribas, A. & Schumacher, T. N. CANCER IMMUNOLOGY. The “cancer immunogram”. Science 352, 658–660 (2016).
Lantuejoul, S. et al. PD-L1 testing for lung cancer in 2019: perspective from the IASLC pathology committee. J. Thorac. Oncol. 15, 499–519 (2020).
Tsao, M. S. et al. PD-L1 immunohistochemistry comparability study in real-life clinical samples: results of blueprint phase 2 project. J. Thorac. Oncol. 13, 1302–1311 (2018).
McLaughlin, J. et al. Quantitative assessment of the heterogeneity of PD-L1 expression in non-small-cell lung cancer. JAMA Oncol. 2, 46–54 (2016).
Ilie, M., Hofman, V., Dietel, M., Soria, J. C. & Hofman, P. Assessment of the PD-L1 status by immunohistochemistry: challenges and perspectives for therapeutic strategies in lung cancer patients. Virchows Arch 468, 511–525 (2016).
Cooper, W. A. et al. Intra- and interobserver reproducibility assessment of PD-L1 biomarker in non-small cell lung cancer. Clin. Cancer Res. 23, 4569–4577 (2017).
Brunnstrom, H. et al. PD-L1 immunohistochemistry in clinical diagnostics of lung cancer: inter-pathologist variability is higher than assay variability. Mod. Pathol. 30, 1411–1421 (2017).
Troncone, G. & Gridelli, C. The reproducibility of PD-L1 scoring in lung cancer: can the pathologists do better? Transl. Lung Cancer Res. 6, S74–S77 (2017).
Niazi, M. K. K., Parwani, A. V. & Gurcan, M. N. Digital pathology and artificial intelligence. Lancet Oncol. 20, e253–e261 (2019).
Tizhoosh, H. R. & Pantanowitz, L. Artificial intelligence and digital pathology: challenges and opportunities. J. Pathol. Inf. 9, 38 (2018).
Salto-Tellez, M., Maxwell, P. & Hamilton, P. Artificial intelligence-the third revolution in pathology. Histopathology 74, 372–376 (2019).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Litjens, G. et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep. 6, 26286 (2016).
Sharma, H., Zerbe, N., Klempert, I., Hellwich, O. & Hufnagl, P. Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology. Comput. Med. Imaging Graph 61, 2–13 (2017).
Ehteshami Bejnordi, B. et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318, 2199–2210 (2017).
Sirinukunwattana, K. et al. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans. Med. Imaging 35, 1196–1206 (2016).
Coudray, N. et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 24, 1559–1567 (2018).
Kather, J. N. et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 25, 1054–1056 (2019).
Nagpal, K. et al. Development and validation of a deep learning algorithm for gleason grading of prostate cancer from biopsy specimens. JAMA Oncol. 6, 1372–1380 (2020).
Mobadersany, P. et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl Acad. Sci. USA 115, E2970–E2979 (2018).
Hamilton, P. W. et al. Digital pathology and image analysis in tissue biomarker research. Methods 70, 59–73 (2014).
Koelzer, V. H., Sirinukunwattana, K., Rittscher, J. & Mertz, K. D. Precision immunoprofiling by image analysis and artificial intelligence. Virchows Arch 474, 511–522 (2019).
Qaiser, T. et al. HER2 challenge contest: a detailed assessment of automated HER2 scoring algorithms in whole slide images of breast cancer tissues. Histopathology 72, 227–238 (2018).
Koelzer, V. H. et al. Digital image analysis improves precision of PD-L1 scoring in cutaneous melanoma. Histopathology 73, 397–406 (2018).
Kapil, A. et al. Deep semi supervised generative learning for automated tumor proportion scoring on NSCLC tissue needle biopsies. Sci. Rep. 8, 17343 (2018).
Taylor, C. R. et al. A multi-institutional study to evaluate automated whole slide scoring of immunohistochemistry for assessment of Programmed Death-Ligand 1 (PD-L1) expression in non-small cell lung cancer. Appl. Immunohistochem. Mol. Morphol. 27, 263–269 (2019).
Bankhead, P. et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
Chang, S., Park, H. K., Choi, Y. L. & Jang, S. J. Interobserver reproducibility of PD-L1 biomarker in non-small cell lung cancer: a multi-institutional study by 27 pathologists. J. Pathol. Transl. Med. 53, 347–353 (2019).
Hirsch, F. R. et al. PD-L1 immunohistochemistry assays for lung cancer: results from phase 1 of the blueprint PD-L1 IHC assay comparison project. J. Thorac. Oncol. 12, 208–222 (2017).
Munari, E. et al. PD-L1 assays 22C3 and SP263 are not interchangeable in non-small cell lung cancer when considering clinically relevant cutoffs: an interclone evaluation by differently trained pathologists. Am. J. Surg. Pathol. 42, 1384–1389 (2018).
Steiner, D. F. et al. Impact of deep learning assistance on the histopathologic review of lymph nodes for metastatic breast cancer. Am. J. Surg. Pathol. 42, 1636–1646 (2018).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82003155 and 81871860), Capital Funds for Health Improvement and Research (2020-2-1025), and the Innovation Fund for Outstanding Doctoral Candidates of Peking University Health Science Center (JH W).
Author information
Authors and Affiliations
Contributions
J. W. and W. S. contributed to image annotations, PD-L1 scoring, manuscript writing, and participated in the experimental design. C. L., X. L., and J. Z. contributed to the DL model, AI system development, manuscript writing, and participated in the experimental design. N. G. and L. L. performed statistical analysis and graphics. Y. Z., H. W., and X. L. contributed to image annotations. X. Y., Y. Z., R. C., and L. M. contributed to PD-L1 scoring. L. Z., K. C., and X. H. performed immunohistochemistry of PD-L1 and prepared the slides. S. L. and D. L. conceived the study, participated in its design and coordination, and helped draft and edit the manuscript. D. L. also verified PD-L1 scoring.
Corresponding authors
Ethics declarations
competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approved by the Peking University Cancer Hospital (PKUCH, No. 2018KT94) and Tianjin Medical University Cancer Hospital (TMUCH, No. Ek2020140). Informed consent was waived by the institutional review boards, because the retrospective archival materials were anonymized, and the results did not have any impact on patient management.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Wu, J., Liu, C., Liu, X. et al. Artificial intelligence-assisted system for precision diagnosis of PD-L1 expression in non-small cell lung cancer. Mod Pathol 35, 403–411 (2022). https://doi.org/10.1038/s41379-021-00904-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-021-00904-9
This article is cited by
-
Artificial intelligence-based assessment of PD-L1 expression in diffuse large B cell lymphoma
npj Precision Oncology (2024)
-
Comparing deep learning and pathologist quantification of cell-level PD-L1 expression in non-small cell lung cancer whole-slide images
Scientific Reports (2024)