Abstract
TP53 mutations drive colorectal cancer development, with missense mutations frequently leading to accumulation of abnormal TP53 protein. TP53 alterations have been associated with poor prognosis and chemotherapy resistance, but data remain controversial. Here, we examined the predictive utility of TP53 overexpression in the context of current adjuvant treatment practice for patients with stage III colorectal cancer. A prospective cohort of 264 stage III patients was tested for association of TP53 expression with 5-year disease-free survival, grouped by adjuvant treatment. Findings were validated in an independent retrospective cohort of 274 stage III patients. Overexpression of TP53 protein (TP53+) was found in 53% and 52% of cases from the prospective and retrospective cohorts, respectively. Among patients receiving adjuvant chemotherapy, TP53+ status was associated with shorter disease-free survival (p ≤ 0.026 for both cohorts), while no difference in outcomes between TP53+ and TP53− cases was observed for patients treated with surgery alone. Considering patients with TP53− tumors, those receiving adjuvant treatment had better outcomes compared with those treated with surgery alone (p ≤ 0.018 for both cohorts), while no treatment benefit was apparent for patients with TP53+ tumors. Combined cohort-stratified analysis adjusted for clinicopathological variables and DNA mismatch repair status confirmed a significant interaction between TP53 expression and adjuvant treatment for disease-free survival (pinteraction = 0.030). For the combined cohort, the multivariate hazard ratio for TP53 overexpression among patients receiving adjuvant chemotherapy was 2.03 (95% confidence interval 1.41–2.95, p < 0.001), while the hazard ratio for adjuvant treatment among patients with TP53− tumors was 0.42 (95% confidence interval 0.24–0.71, p = 0.001). Findings were maintained irrespective of tumor location or when restricted to mismatch repair-proficient tumors. Our data suggest that adjuvant chemotherapy benefit in stage III colorectal cancer is restricted to cases with low-level TP53 protein expression. Identifying TP53+ tumors could highlight patients that may benefit from more aggressive treatment or follow-up.
Similar content being viewed by others
Introduction
5-Fluorouracil-based adjuvant chemotherapy is the standard-of-care for patients with stage III colorectal cancer [1, 2], although some individuals are too frail or decline treatment. Despite being the mainstay of care, the benefit from adjuvant chemotherapy is limited to only 10–15% of individuals and 5-year relapse rates remain at ~60% [3]. Overtreatment with adjuvant chemotherapy (85–90% of patients) is associated with significant toxicities and healthcare costs. As a result, considerable efforts have been invested in the identification of tumor-based molecular markers that can complement standard clinical and pathological staging systems to more accurately predict disease outcome and determine optimal adjuvant treatment approaches. Whilst several markers associated with prognosis have been identified in colorectal cancer, such as DNA mismatch repair status and gene expression profiles [4, 5], there remains a clinical need for biomarkers predictive of which patients will derive a benefit from treatment with chemotherapy.
Mutation of the TP53 tumor suppressor is a central driver of colorectal carcinogenesis found in ~50% of sporadic tumors [6]. Wild-type TP53 protein mediates cell-cycle arrest and cell-death checkpoints, which can be triggered by various cellular stress signals [7]. Normal cells under unperturbed conditions express TP53 at low levels, with its degradation mediated by ubiquitin-ligases including MDM2, COP-1, and PIRH-2 [8,9,10]. In most tumors, both TP53 alleles are inactivated, usually by a combination of a missense mutation and 17p deletion that eliminates the second TP53 allele [11]. TP53 missense mutations frequently lead to accumulation of abnormal TP53 protein with a prolonged half-life in the nucleus, which can be detected by immunohistochemistry [12, 13]. Studies that have examined both TP53 expression and mutation data for colorectal cancer have reported an agreement of immunohistochemistry and mutation detection between 53 and 76% [14,15,16,17,18,19,20].
TP53 aberrations are considered a late event in the classic adenoma-to-carcinoma sequence of colorectal tumorigenesis, associated with the transition from adenoma to carcinoma [21]. Some evidence suggest that the frequency of TP53 aberrations increases with tumor stage [22,23,24,25,26,27,28,29,30,31], although this has not been confirmed in other studies [16, 28, 32,33,34,35,36,37,38,39,40]. There is general agreement that TP53 aberrations occur less frequently in the proximal colon as compared with the distal colon and rectum [18,19,20, 30, 34, 39, 41,42,43,44,45,46,47,48].
The significance of TP53 aberrations as a prognostic marker for colorectal cancer remains a matter of controversy. For investigations of TP53 protein expression which included at least 100 patients, overexpression of TP53 has been associated with inferior outcomes in univariate or multivariate survival analyses in multiple reports [19, 20, 26,27,28, 31,32,33, 35, 39, 40, 49,50,51,52,53,54,55,56,57,58]. However, other studies have found no association [18, 22, 25, 30, 37, 41, 45, 46, 48, 59,60,61,62,63,64,65] or reported the opposite finding [34, 38, 66,67,68]. A systematic review of TP53 expression data performed by Munro et al. some time ago, which encompassed data for 12,257 patients across all tumor stages, concluded that individuals with abnormal TP53 were at increased risk of death (relative risk 1.32, 95% confidence interval 1.23–1.42), with similar results for assessment of mutation data (relative risk 1.31, 95% confidence interval 1.19–1.45) [69]. However, both publication bias and heterogeneity of results were noted.
The lack of a consensus in the literature on the prognostic significance of TP53 aberrations may be due to the use of heterogeneous study populations such as tumor stages included, differences in immunohistochemical methods, limited cohort sizes and duration of follow-up. A major issue is absence of a standardized immunohistochemistry scoring scheme to optimally correlate TP53 protein expression with TP53 mutation status. It has further been suggested that the prognostic significance of TP53 aberrations may depend on the ethnic group, body-mass index, tumor location, or stage of disease [34, 39, 42, 43, 52]. TP53 aberrations are also negatively associated with tumor microsatellite instability status [20, 39, 44, 70,71,72], found in ~15% of colorectal cancers, an established marker of good prognosis for early-stage colorectal cancer [4]. Microsatellite instability is characterized by increased insertion or deletion mutations at simple repeat sequences due to a defect in DNA mismatch repair [73], and may be associated with a lack of benefit from 5-fluorouracil-based adjuvant chemotherapy [74,75,76,77].
Furthermore, there are data from colorectal cancer cell lines studies to suggest that the benefit of adjuvant chemotherapy may be limited to patients with TP53 wild-type cancers. Specifically, colorectal cancer cells with wild-type TP53 have consistently been found to be more sensitive to 5-fluorouracil and oxaliplatin treatment as compared with cells with mutated TP53 [78,79,80,81,82,83]. Accordingly, some patient cohort studies have reported inferior outcomes for individuals receiving chemotherapy if their tumors had TP53 overexpression [20, 38, 84, 85], but this was not observed by others [30, 55, 86,87,88]. The previous systematic review of Munro at el. did not find a relationship between TP53 aberrations and adjuvant chemotherapy benefit, although it identified a relationship with neoadjuvant chemoradiation for rectal cancer [69]. Results of the TP53 Colorectal Cancer International Collaborative Study, which combined TP53 mutation data from 25 different research groups in 17 countries, indicated a potential interaction between TP53 mutation status and adjuvant treatment benefit for Dukes’ C patients with distal tumors [89].
To clarify the conflicting data regarding the prognostic and predictive value of TP53 protein expression in early-stage colorectal cancer, we evaluated TP53 expression in a homogenous prospective community-based population of 264 stage III patients. Specifically, the present study sought to clarify the clinical potential of TP53 overexpression as a predictor of outcomes in the context of current adjuvant treatment practice. An optimal cutoff for identifying TP53 overexpression (TP53+ vs TP53−) was determined using the Allred scoring system [90], which considers both proportion of stained cells and stain intensity, based on a set of 66 colorectal cancer cell lines with existing TP53 mutation data. Findings were validated in an independent retrospective community-based cohort of 274 stage III patients. Hazard ratios and recurrence rates were estimated for patient subgroups by TP53 expression status and adjuvant treatment for a cohort-stratified analysis of the combined 538 patient population.
Materials and methods
Prospective community series of colorectal cancer patients
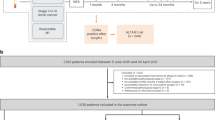
A total of 264 patients with resected stage III colorectal adenocarcinoma were recruited at the Royal Melbourne Hospital and Western Hospital, Footscray in Australia between 1999 and 2011. Individuals with hereditary polyposis colorectal cancer syndromes or who had received neoadjuvant chemoradiation were excluded. Formalin-fixed paraffin-embedded tumor and matched normal tissue specimens were obtained at surgery, and tissue microarrays consisting of 1-mm-diameter tissue cores were constructed. For each patient, four tumor and two normal tissue cores were embedded, with tumor cores taken from areas of the densest tumor cell percentage. A total of 189 (72%) patients received 5-fluorouracil-based adjuvant chemotherapy. All patients were prospectively followed according to the standard protocols, with 3 monthly clinic visits and testing for carcinoembryonic antigen levels, 12 monthly CT scans of the chest, abdomen and pelvis for 2 years after diagnosis and then 6 monthly clinic visits and carcinoembryonic antigen testing until 5 years from diagnosis. Patient characteristics are summarized in Supplementary Table S1. All participants gave informed consent, and this study was approved by the medical ethics committees of all sites (HREC 12/19).
TP53 protein expression
The DO-7 mouse monoclonal anti-TP53 antibody (Novocastra) was used for immunohistochemistry. Tissue staining was performed on tissue microarray sections as per standard protocol on a BenchMark ULTRA platform (Ventana). Briefly, heat-induced antigen retrieval was performed using CC1 (EDTA) buffer at 95 °C for 36 min. Tissue sections were incubated at 36 °C with the primary TP53 antibody at a 1:100 dilution for 32 min, followed by signal detection using an enzyme-conjugated multimer secondary antibody (UltraView Universal DAB detection kit, Ventana). Sections were counterstained with hematoxylin.
Evaluation of TP53 staining was carried out by a gastrointestinal pathologist (DSW) blinded to all clinical information. We considered only tumor cells with distinct nuclear immunostaining for TP53 as positive (Supplementary Fig. S1). Considering all tumor cores, the Allred score was calculated by adding a score reflecting the percentage of positive tumor cells (0 for none, 1 for <1%, 2 for 1–10%, 3 for 11–33%, 4 for 34–66%, and 5 for 67–100%) and a score reflecting the intensity of immunoreactivity (1 for weak, 2 for moderate, and 3 for strong), with a maximum score of 8 [90]. Based on a set of colorectal cancer cell lines (n = 66) with known TP53 mutation status [91], an Allred score of 6 or greater was selected, reflecting the cutoff value identifying TP53 missense mutated cases with maximum sensitivity and specificity (Supplementary Methods, Supplementary Fig. S2). To assess the accuracy of our colorectal cancer cell line determined cut-off in primary tumor tissue, we integrated immunohistochemistry scores with TP53 mutation data available from Sanger sequencing on a subset of 51 tumor samples [92]. A total of 25 of 26 (96%) samples with TP53 missense mutation exhibited an Allred score of >6, while 19 of 25 (76%) samples without TP53 missense mutation exhibited an Allred score of ≤6. Reproducibility of TP53 expression scores was verified by re-examination of a random selection of 454 cases across both our study cohorts by a second observer (CB), blinded to all prior data. Interobserver reproducibility for TP53 scoring showed high concordance (κ statistic = 0.85; 95% confidence interval = 0.80–0.90).
DNA mismatch repair status
Whole-tissue sections were cut from tumor and matched normal formalin-fixed paraffin-embedded blocks. For tumor samples, macrodissection was performed to enrich for areas with >60% neoplastic cells guided by hematoxylin and eosin stained tissue sections. DNA was extracted using standard protocols and polymerase chain reaction amplified for the Bethesda consensus panel of microsatellite markers (BAT25, BAT26, D2S123, D5S346, and D17S250) using fluorescently labeled primers [93]. Polymerase chain reaction products were analyzed on a 3130xl Genetic Analyzer (Applied Biosystems). Mismatch repair-deficient status was diagnosed if instability was detected at two or more markers, and mismatch repair-proficient status was diagnosed if instability was detected at fewer than two markers.
Retrospective community series of colorectal cancer patients
To validate findings, we examined an independent cohort of 274 patients with stage III colorectal cancer recruited at Austin Health, Australia, between 1998 and 2015, for whom clinical, treatment and follow-up data were retrospectively assembled with human research ethics approval (HREC H2013/05077). Among these patients, 181 (66%) had received 5-fluorouracil-based adjuvant chemotherapy. Individuals with hereditary polyposis colorectal cancer syndromes or who had received neoadjuvant chemoradiation were excluded. Patient characteristics are summarized in Supplementary Table S1. Archival formalin-fixed paraffin-embedded tumor blocks were retrieved, and tissue microarrays prepared with sampling of three 1-mm-diameter cores per patient. Immunohistochemistry for TP53 expression was performed on the same platform as for the prospective community series. For this cohort, tumor DNA mismatch repair status was assigned based on standard immunohistochemistry diagnostic assays for mismatch repair proteins (MLH1, MSH2, PMS2, and MSH6) by pathologist DSW [94]. Mismatch repair-deficient was defined as absence of nuclear staining in tumor cells but positive nuclear staining in stromal cells and lymphocytes for at least one mismatch repair protein.
Statistical methods
Statistical analyses were conducted using the statistical computing software R (R Development Core Team, 2011). Receiver operator curves were used to determine the optimal Allred score cutoff for TP53 immunohistochemistry to detect TP53 missense mutations (see Supplementary Methods). Interobserver reproducibility for determination of TP53 expression status between reviewers was assessed using Cohen’s kappa statistic. For univariate analyses, differences between groups were assessed using the Fisher’s exact test for categorical variables and the Kruskal–Wallis test for continuous variables. Outcome analyses were conducted for 5-year disease-free survival. Disease-free survival was defined as time from surgery to the first confirmed relapse, with censoring done when a patient died or was alive without recurrence at last contact. Survival curves were generated according to the method of Kaplan and Meier. Cox proportional hazard models were used to assess the associations of TP53 expression status with disease-free survival in the context of patient characteristics, DNA mismatch repair status and adjuvant treatment. Hazard ratios and 95% confidence intervals were calculated. All statistical analyses were two-sided and considered significant if p < 0.050.
Results
Patient characteristics of the prospective community-based cohort
Of the 264 patients with stage III adenocarcinoma, 102 (39%) were female, and the median patient age was 70 (range 39–91) years (Supplementary Table S1). A total of 106 (40%) cancers were from the proximal colon and 158 (60%) from the distal colon or rectum; 168 (64%) exhibited low and 96 (36%) high grade. All patients underwent surgical resection, and 189 (72%) patients received 5-fluorouracil-based adjuvant chemotherapy. A total of 91 (34%) patients experienced a disease recurrence. The median follow-up duration was 26.6 (range 3.4–60.0) months for individuals with recurrence and 60.0 (range 0.3–60.0) months for individuals without recurrence.
Overexpression of TP53 was scored based on the intensity and percentage of stained tumor nuclei using a pre-determined Allred score cutoff of ≥6, optimized to identify TP53 missense mutations in a set of 66 colorectal cancer cell lines (Supplementary Methods, Supplementary Figs. S1 and S2). Nonneoplastic colonic mucosa, inflammatory, and stromal cells exhibited weak staining and served as positive internal controls. Overall, 53% (140 of 264) of the tumors exhibited TP53 overexpression, termed TP53+. For mismatch repair status 20% (52 of 264) of the tumors were mismatch repair deficient.
Compared with TP53− tumors, the TP53+ tumors were associated with distal location (odds ratio 1.7, 95% confidence interval 1.0–2.9, p = 0.044) and mismatch repair-proficient status (odds ratio 2.1, 95% confidence interval 1.1–4.1, p = 0.021) (Table 1). There was no association of TP53 expression status with gender, age at presentation or grade. Mismatch repair-deficient status was positively correlated with proximal tumor location (odds ratio 6.4, 95% confidence interval 3.1–14.1, p < 0.001) and high grade (odds ratio 3.0, 95% confidence interval 1.6–5.9, p < 0.001).
TP53 expression status and disease-free survival
Overall, patients with TP53+ tumors had inferior 5-year disease-free survival rates than patients with TP53− tumors (54.5% vs 68.9%). The univariate hazard ratio for disease-free survival for the TP53+ group versus the TP53− group was 1.64 (95% confidence interval 1.08–2.52; p = 0.020). In addition, proximal tumor location and high grade were associated with reduced disease-free survival while adjuvant treatment was associated with improved disease-free survival (p < 0.020 for all univariate analyses). Patient gender, age at diagnosis, and mismatch repair status were not associated with outcome in our cohort. In multivariate analysis adjusting for clinicopathological variables significant in the univariate models, TP53 expression remained an independent predictor of poor prognosis (hazard ratio 1.93, 95% confidence interval 1.25–2.97, p = 0.003).
Adjuvant treatment benefit by TP53 expression status
We next examined whether tumor TP53 expression status was associated with differential benefit from adjuvant treatment. Notably, patients who had received adjuvant treatment were significantly younger than individuals who had not received adjuvant treatment (66 vs 82 years; p < 0.001); this trend was similar across the TP53 subgroups (data not shown). The duration of follow-up according to adjuvant treatment was similar between TP53 subgroups (p > 0.066).
Considering patients grouped by adjuvant treatment, association of TP53 expression status with outcome appeared restricted to individuals receiving chemotherapy. Among adjuvant therapy treated patients, TP53+ tumors showed significantly poorer outcomes than TP53− tumors with a univariate hazard ratio for disease-free survival of 2.09 (95% confidence interval 1.23–3.56, p = 0.005) and 5-year disease-free survival rates of 55.7% (95% confidence interval 46.5–66.7%) and 76.3% (95% confidence interval 67.8–86.0%), respectively (Fig. 1a). In multivariate analysis adjusting for clinicopathological variables significant in univariate analysis, the hazard ratio for TP53+ tumors in the adjuvant treatment group was 2.59 (95% confidence interval 1.50–4.47; p < 0.001). No difference in outcome by TP53 expression status was apparent for patients treated with surgery alone (univariate hazard ratio 1.02, 95% confidence interval 0.49–2.12, p = 0.956, Fig. 1b).
Kaplan–Meier plot for disease-free survival in patients with stage III colorectal cancer according to TP53 expression status for a individuals treated with adjuvant therapy from the prospective cohort (n = 189), b individuals treated with surgery alone from the prospective cohort (n = 75), c individuals treated with adjuvant therapy from the retrospective cohort (n = 181), and d individuals treated with surgery alone from the retrospective cohort (n = 93)
For groups of patients by TP53 expression status, adjuvant treatment benefit was evident for TP53− tumors, but not for TP53+ tumors. Among patients with TP53− tumors, the hazard ratio for disease-free survival for adjuvant treatment as compared with surgery alone, adjusting for variables significant in univariate models, was 0.41 (95% confidence interval 0.19–0.75; p = 0.008) (Fig. 2a). In contrast, among TP53+ patients, the univariate hazard ratio for disease-free survival for adjuvant treatment was 0.79 (95% confidence interval, 0.44–1.42; p = 0.434) (Fig. 2b).
Kaplan–Meier plot for disease-free survival in patients with stage III colorectal cancer according to adjuvant treatment for a individuals with TP53− tumors from the prospective cohort (n = 124), b individuals with TP53+ tumors from the prospective cohort (n = 140), c individuals with TP53− tumors from the retrospective cohort (n = 132), and d individuals with TP53+ tumors from the retrospective cohort (n = 142)
Independent retrospective community cohort
We validated the differential outcome associations for TP53 expression by adjuvant treatment in an independent retrospective population of 274 patients with stage III colorectal cancer, 66% (181 of 274) of whom had received adjuvant chemotherapy (Supplementary Table S1). A total of 108 (39%) of patients in this cohort had experienced relapse. The median follow-up duration was 36.0 (range, 4.3–60.0) months for individuals with recurrence and 59.0 (range, 0.7–60.0) months for individuals without recurrence. Patients in this cohort showed a similar proportion of TP53+ tumors (52%, 142 of 274) as compared with our prospective cohort; again, TP53 overexpression was inversely associated with mismatch repair-deficient status and proximal tumor location (p < 0.001 for both comparisons, Table 1).
Outcomes among the treatment groups mirrored the findings from the prospective cohort: TP53+ tumors exhibited inferior disease-free survival than TP53− tumors for patients receiving adjuvant chemotherapy (multivariate hazard ratio 1.73, 95% confidence interval 1.07–2.79, p = 0.026, Fig. 1c), while similar outcomes were observed for patients treated with surgery alone (multivariate hazard ratio 0.94, 95% confidence interval 0.48–1.82, p = 0.847, Fig. 1d). Again, adjuvant treatment benefit appeared limited to patients with TP53− tumors (multivariate hazard ratio 0.48, 95% confidence interval 0.27–0.88, p = 0.018, Fig. 2c), with no apparent benefit in TP53+ patients (multivariate hazard ratio 1.08, 95% confidence interval 0.61–1.90, p = 0.798, Fig. 2d).
In combined cohort-stratified analysis, multivariate analysis adjusted for all clinicopathological variables and mismatch repair status confirmed a significant interaction between TP53 expression status and adjuvant treatment for disease-free survival (pinteraction = 0.030). The 5-year disease-free survival rate was highest for patients with TP53-/adjuvant treated tumors (71.6%, 95% confidence interval 64.9–78.9%), while disease-free survival rates were significantly lower for patients with TP53+/adjuvant treated tumors (52.9%, 95% confidence interval 46.3–60.6%), patients with TP53+/surgery alone tumors (48.1%, 95% confidence interval 36.1–64.0%) and patients with TP53-/surgery alone tumors (47.5%, 95% confidence interval 36.1–62.7%) (Fig. 3). For the combined cohort, the multivariate hazard ratio for TP53 overexpression among patients receiving adjuvant chemotherapy was 2.03 (95% confidence interval 1.41–2.95, p < 0.001, Table 2), while the multivariate hazard ratio for adjuvant treatment among patients with TP53− tumors was 0.42 (95% confidence interval 0.24–0.71, p = 0.001, Table 3). The relationship between TP53 overexpression and inferior outcome among patients receiving adjuvant chemotherapy was maintained when extending the multivariate analysis to include further available National Comprehensive Cancer Network high-risk features, T4 stage and extramural venous invasion (Supplementary Table S2). Improved survival rates for patients with TP53-/adjuvant treated tumors were found when separately analyzing patients with proximal and distal cancers, or when restricting the analysis to patients with mismatch repair-proficient tumors (Supplementary Table S3).
Discussion
The present study sought to clarify the clinical potential of TP53 overexpression as a predictor of outcomes for patients with stage III colorectal cancer in the context of current adjuvant treatment practice. We examined a total of 538 patients across two independent cohorts drawn from prospective and retrospective community-based patient populations. Among patients receiving adjuvant chemotherapy, TP53+status was associated with shorter disease-free survival for both cohorts, while no difference in outcomes between TP53+ and TP53− cases was observed for patients treated with surgery alone. Considering patients with TP53- tumors, those receiving adjuvant treatment had better outcomes as compared those treated with surgery alone, while no significant benefit from adjuvant treatment was apparent for patients with TP53+ tumors. Combined cohort-stratified analysis adjusted for clinicopathological variables and mismatch repair status confirmed a significant interaction between tumor TP53 expression status and adjuvant treatment for disease-free survival. Taken together, these data are consistent with TP53 overexpression-related resistance to 5-fluorouracil-based adjuvant chemotherapy, highlighting TP53+ patients as a subset to consider for more aggressive treatment or follow-up. Activation of wild-type TP53 plays a pivotal role in triggering apoptosis in response to chemotherapeutic agents, and the predictive value of TP53 overexpression may be related to mutated protein impairing downstream transcriptional activity and abrogating TP53 interaction with pro-survival BCL-2 family proteins [95,96,97,98].
Missense mutations in TP53 are common in sporadic colorectal cancer, and often result in accumulation of TP53 protein. However, there is presently no standardized clinical scoring system for evaluating TP53 status by immunohistochemistry. Studies using the DO-7 antibody report TP53 overexpression in between 30 and 63% of colorectal cancers [18,19,20, 22, 30, 34,35,36, 38, 42, 45, 47, 54, 57, 61, 64, 65, 88, 99]. Using a separate training cohort of 66 colorectal cancer cell lines with known TP53 mutation status, we determined that an Allred score of ≥6, which considers both proportion of stained cells and stain intensity, optimally identified cases with TP53 missense mutations. Applying this cutoff to our prospective and retrospective patient cohorts, we identified TP53 overexpression in 53% and 52% of cases, respectively. Multiple reports have discussed the relationship of TP53 aberrations with location of the tumor [18,19,20, 30, 34, 39, 41,42,43,44,45,46,47,48]. Consistent with these studies, we found that the frequency of TP53 overexpression was reduced in tumors from the proximal colon as compared with the distal colon and rectum. We also observed the well-established negative association between TP53 aberrations and mismatch repair deficiency [39, 44, 70,71,72].
Data regarding the prognostic role of TP53 expression in colorectal cancer are heterogeneous. Several studies have reported that TP53 overexpression is an adverse prognostic factor [19, 20, 26,27,28, 31,32,33, 35, 39, 40, 49,50,51,52,53,54,55,56,57,58], and this overall trend was evident in our study for combined cohort-stratified multivariate analysis with baseline variables (Supplementary Table S4) and with additional inclusion of T4 stage and extramural venous invasion (Supplementary Table S5). Nonetheless, other studies found no relationship with outcome [18, 22, 25, 30, 37, 41, 45, 46, 48, 59,60,61,62,63,64,65] and a few reports observed the opposite association [34, 38, 66,67,68].
Besides differences in TP53 assay methodologies, scoring schemes, cohort heterogeneity, study sizes and duration of follow-up, in vitro studies have highlighted TP53 aberration-associated resistance to 5-fluorouracil and oxaliplatin as a potential major confounding factor of colorectal cancer prognostic studies [78,79,80,81,82,83]. In agreement with these observations, we found that for patients receiving adjuvant chemotherapy, individuals with TP53+ tumors had significantly poorer outcomes than individuals with TP53− tumors. For patients grouped by TP53 expression status, adjuvant treatment was associated with improved outcomes for individuals with TP53− tumors, but not for individuals with TP53+ tumors. Improved survival rates for patients with TP53-/adjuvant treated tumors were observed irrespective of tumor location and when restricting the analysis to patients with mismatch repair-proficient tumors. However, these results should be interpreted with caution because of the non-randomized use of adjuvant chemotherapy. In addition, we considered all FU-based treatment regimens as one group, although TP53 aberrations may show different predictive values according to the exact type of treatment used [47]. Bearing in mind these caveats, our results support the contention that the benefit from adjuvant chemotherapy depends on TP53 expression status. Consistent with our data, several cohort studies have reported an apparent lack of benefit from chemotherapy for early-stage and metastatic tumors with TP53 overexpression [20, 38, 84, 85]. However, other reports of patients with early-stage colorectal cancer did not reproduce this observation [30, 55, 86,87,88], although these differed in methods or cut-offs for scoring TP53 status and included patients with stage II tumors. These studies lacked validation cohorts, and in one study inspection of the Kaplan–Meier curves shows a trend towards chemotherapy benefit in the TP53-ve group [87]. Another study of adjuvant chemotherapy treated patients identified poorer outcomes for TP53+ tumors only if these were also high for BAX expression [72].
In our study, we ensured technical quality by performing the immunohistochemistry detection in an accredited diagnostic laboratory, using the Allred scoring system with a pre-determined TP53 overexpression cutoff optimized to detect TP53 missense mutations. Reproducibility of TP53 immune-staining scores was confirmed by two reviewers blinded to all clinical data. Clinicopathologic and outcome associations with TP53 status were validated across two independent cohorts. Immunohistochemistry is an attractive approach for detecting biomarkers due to its applicability to routine archival clinical specimens, relatively low cost, rapid test turn-around time, straightforward methodology and analysis. Immunohistochemistry can be readily incorporated into existing panels of biomarkers for specific cancer subtypes.
Limitations of this study include that our cohorts were derived from population-based series treated according to standard-practice for adjuvant therapy, not a randomized clinical trial. As a result of this study design, there is potential bias for poorer outcomes in the untreated population group, likely to be older, frailer or subject to more comorbidities. For this reason, disease-free survival was chosen as the preferred measure for response to therapy rather than overall survival, since disease-free survival is less likely to be impacted by age, frailty, or comorbidities. The study was not standardized for adjuvant treatment regimen, with single-agent 5-fluorouracil and 5-fluorouracil/oxaliplatin combination treatments used as per routine care. As stage III patients cannot ethically be randomized to adjuvant chemotherapy versus surgery alone, analysis of tumor samples from previous randomized clinical trials would be the most appropriate approach for validation of our findings and further examination of TP53 predictive value by treatment regimen. Another limitation of our study is that TP53 immunohistochemistry assessment was based on tissue microarray cores, which may not be representative of the entire tumor due to heterogeneity. Sampling of at least three cores from different regions of tumor should have addressed this issue to some extent. Consistent with most previous reports, our study focused on scoring TP53 overexpression, associated with missense mutations; this excludes loss events due to nonsense, frameshift or splice-site mutations (bi-allelic or with loss of heterozygosity) which account for a subset of ~6% of TP53 mutated colorectal cancers as estimated from The Cancer Genome Atlas whole-exome sequencing data on colorectal cancers [11]. Conversely, posttranscriptional stabilization processes such as those induced by DNA damage can lead to accumulation of wild-type TP53 protein [100].
In conclusion, our examination of the predictive value of TP53 expression in the context of current adjuvant treatment practice suggests that overexpression of TP53 protein is associated with minimal adjuvant chemotherapy benefit in patients with stage III colorectal cancer. Notably the disease-free survival rate for patients with TP53+ tumors receiving adjuvant treatment was similar to the disease-free survival rates in patients with TP53+ or TP53− tumors treated with surgery alone. Our findings indicate patients with TP53+ tumors as a subset to consider for more aggressive treatment or follow-up. Further evaluation of TP53 expression status as a predictive biomarker for adjuvant therapy appears warranted, utilizing tumor samples from previous randomized clinical trials to determine an optimal diagnostic approach using standardized immunohistochemistry scoring methods and comparison with sequencing-based testing. Allred scores of 7–8 have very high specificity for TP53 mutation, and immunohistochemistry can serve as a cost-effective primary screen to identify these cases. Sequencing might optimally be used to identify non-responders to chemotherapy in equivocal (Allred 6) cases, or for tumors lacking TP53 expression which may harbor inactivating TP53 mutations.
References
Kerr DJ, Gray R, McConkey C, Barnwell J. Adjuvant chemotherapy with 5-fluorouracil, l-folinic acid and levamisole for patients with colorectal cancer: non-randomised comparison of weekly versus four-weekly schedules—less pain, same gain. QUASAR Colorectal Cancer Study Group. Ann Oncol. 2000;11:947–55.
Andre T, Boni C, Mounedji-Boudiaf L, Navarro M, Tabernero J, Hickish T, et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med. 2004;350:2343–51.
Bockelman C, Engelmann BE, Kaprio T, Hansen TF, Glimelius B. Risk of recurrence in patients with colon cancer stage II and III: a systematic review and meta-analysis of recent literature. Acta Oncol. 2015;54:5–16.
Guastadisegni C, Colafranceschi M, Ottini L, Dogliotti E. Microsatellite instability as a marker of prognosis and response to therapy: a meta-analysis of colorectal cancer survival data. Eur J Cancer. 2010;46:2788–98.
Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–6.
Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217–21.
Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–70.
Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–45.
Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–91.
Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92.
Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7.
Finlay CA, Hinds PW, Tan TH, Eliyahu D, Oren M, Levine AJ. Activating mutations for transformation by p53 produce a gene product that forms an hsc70-p53 complex with an altered half-life. Mol Cell Biol. 1988;8:531–9.
Rodrigues NR, Rowan A, Smith ME, Kerr IB, Bodmer WF, Gannon JV, et al. p53 mutations in colorectal cancer. Proc Natl Acad Sci USA. 1990;87:7555–9.
Smith DR, Ji CY, Goh HS. Prognostic significance of p53 overexpression and mutation in colorectal adenocarcinomas. Br J Cancer. 1996;74:216–23.
Soong R, Robbins PD, Dix BR, Grieu F, Lim B, Knowles S, et al. Concordance between p53 protein overexpression and gene mutation in a large series of common human carcinomas. Hum Pathol. 1996;27:1050–5.
Leahy DT, Salman R, Mulcahy H, Sheahan K, O’Donoghue DP, Parfrey NA. Prognostic significance of p53 abnormalities in colorectal carcinoma detected by PCR-SSCP and immunohistochemical analysis. J Pathol. 1996;180:364–70.
Caldes T, Iniesta P, Vega FJ, de Juan C, Lopez JA, Diaz-Rubio E, et al. Comparative survival analysis of p53 gene mutations and protein accumulation in colorectal cancer. Oncology. 1998;55:249–57.
Kressner U, Inganas M, Byding S, Blikstad I, Pahlman L, Glimelius B, et al. Prognostic value of p53 genetic changes in colorectal cancer. J Clin Oncol. 1999;17:593–9.
Bouzourene H, Gervaz P, Cerottini JP, Benhattar J, Chaubert P, Saraga E, et al. p53 and Ki-ras as prognostic factors for Dukes’ stage B colorectal cancer. Eur J Cancer. 2000;36:1008–15.
Elsaleh H, Powell B, McCaul K, Grieu F, Grant R, Joseph D, et al. P53 alteration and microsatellite instability have predictive value for survival benefit from chemotherapy in stage III colorectal carcinoma. Clin Cancer Res. 2001;7:1343–9.
Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67.
Mulder JW, Baas IO, Polak MM, Goodman SN, Offerhaus GJ. Evaluation of p53 protein expression as a marker for long-term prognosis in colorectal carcinoma. Br J Cancer. 1995;71:1257–62.
Kastrinakis WV, Ramchurren N, Rieger KM, Hess DT, Loda M, Steele G, et al. Increased incidence of p53 mutations is associated with hepatic metastasis in colorectal neoplastic progression. Oncogene. 1995;11:647–52.
Flamini G, Curigliano G, Ratto C, Astone A, Ferretti G, Nucera P, et al. Prognostic significance of cytoplasmic p53 overexpression in colorectal cancer. Immunohistochem Anal Eur J Cancer. 1996;32A:802–6.
Theodoropoulos GE, Karafoka E, Papailiou JG, Stamopoulos P, Zambirinis CP, Bramis K, et al. P53 and EGFR expression in colorectal cancer: a reappraisal of ‘old’ tissue markers in patients with long follow-up. Anticancer Res. 2009;29:785–91.
Pancione M, Forte N, Fucci A, Sabatino L, Febbraro A, Di Blasi A, et al. Prognostic role of beta-catenin and p53 expression in the metastatic progression of sporadic colorectal cancer. Hum Pathol. 2010;41:867–76.
Chen J, Tang H, Wu Z, Zhou C, Jiang T, Xue Y, et al. Overexpression of RBBP6, alone or combined with mutant TP53, is predictive of poor prognosis in colon cancer. PLoS ONE. 2013;8:e66524.
Zhang M, Cui F, Lu S, Lu H, Jiang T, Chen J, et al. Increased expression of prothymosin-alpha, independently or combined with TP53, correlates with poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7:4867–76.
Ji L, Wei Y, Jiang T, Wang S. Correlation of Nrf2, NQO1, MRP1, cmyc and p53 in colorectal cancer and their relationships to clinicopathologic features and survival. Int J Clin Exp Pathol. 2014;7:1124–31.
McGregor MJ, Fadhil W, Wharton R, Yanagisawa Y, Presz M, Pritchard A, et al. Aberrant P53 expression lacks prognostic or predictive significance in colorectal cancer: results from the VICTOR trial. Anticancer Res. 2015;35:1641–5.
Wang P, Liang J, Wang Z, Hou H, Shi L, Zhou Z. The prognostic value of p53 positive in colorectal cancer: a retrospective cohort study. Tumour Biol. 2017;39:1010428317703651.
Yamaguchi A, Nakagawara G, Kurosaka Y, Nishimura G, Yonemura Y, Miyazaki I. p53 immunoreaction in endoscopic biopsy specimens of colorectal cancer, and its prognostic significance. Br J Cancer. 1993;68:399–402.
Zeng ZS, Sarkis AS, Zhang ZF, Klimstra DS, Charytonowicz E, Guillem JG, et al. p53 nuclear overexpression: an independent predictor of survival in lymph node—positive colorectal cancer patients. J Clin Oncol. 1994;12:2043–50.
Soong R, Grieu F, Robbins P, Dix B, Chen D, Parsons R, et al. p53 alterations are associated with improved prognosis in distal colonic carcinomas. Clin Cancer Res. 1997;3:1405–11.
Maeda K, Chung YS, Kang SM, Ogawa M, Onoda N, Nakata B, et al. Overexpression of cyclin D1 and p53 associated with disease recurrence in colorectal adenocarcinoma. Int J Cancer. 1997;74:310–5.
Tollenaar RA, van Krieken JH, van Slooten HJ, Bruinvels DJ, Nelemans KM, van den Broek LJ, et al. Immunohistochemical detection of p53 and Bcl-2 in colorectal carcinoma: no evidence for prognostic significance. Br J Cancer. 1998;77:1842–7.
Poller DN, Baxter KJ, Shepherd NA. p53 and Rb1 protein expression: are they prognostically useful in colorectal cancer? Br J Cancer. 1997;75:87–93.
Ahnen DJ, Feigl P, Quan G, Fenoglio-Preiser C, Lovato LC, Bunn PA, et al. Ki-ras mutation and p53 overexpression predict the clinical behavior of colorectal cancer: a Southwest Oncology Group Study. Cancer Res. 1998;58:1149–58.
Morikawa T, Kuchiba A, Liao X, Imamura Y, Yamauchi M, Qian ZR, et al. Tumor TP53 expression status, body mass index and prognosis in colorectal cancer. Int J Cancer. 2012;131:1169–78.
Li C, Bu J, Liao Y, Zhang J, Han J, Zhang H, et al. High expressions of CUL4A and TP53 in colorectal cancer predict poor survival. Cell Physiol Biochem. 2018;51:2829–42.
Bell SM, Scott N, Cross D, Sagar P, Lewis FA, Blair GE, et al. Prognostic value of p53 overexpression and c-Ki-ras gene mutations in colorectal cancer. Gastroenterology. 1993;104:57–64.
Gervaz P, Bouzourene H, Cerottini JP, Chaubert P, Benhattar J, Secic M, et al. Dukes B colorectal cancer: distinct genetic categories and clinical outcome based on proximal or distal tumor location. Dis Colon Rectum. 2001;44:364–72. discussion 72-3.
Diez M, Medrano M, Muguerza JM, Ramos P, Hernandez P, Villeta R, et al. Influence of tumor localization on the prognostic value of P53 protein in colorectal adenocarcinomas. Anticancer Res. 2000;20:3907–12.
Samowitz WS, Holden JA, Curtin K, Edwards SL, Walker AR, Lin HA, et al. Inverse relationship between microsatellite instability and K-ras and p53 gene alterations in colon cancer. Am J Pathol. 2001;158:1517–24.
Bazan V, Migliavacca M, Zanna I, Tubiolo C, Corsale S, Calo V, et al. DNA ploidy and S-phase fraction, but not p53 or NM23-H1 expression, predict outcome in colorectal cancer patients. Result of a 5-year prospective study. J Cancer Res Clin Oncol. 2002;128:650–8.
Tornillo L, Lugli A, Zlobec I, Willi N, Glatz K, Lehmann F, et al. Prognostic value of cell cycle and apoptosis regulatory proteins in mismatch repair-proficient colorectal cancer: a tissue microarray-based approach. Am J Clin Pathol. 2007;127:114–23.
Zaanan A, Cuilliere-Dartigues P, Guilloux A, Parc Y, Louvet C, de Gramont A, et al. Impact of p53 expression and microsatellite instability on stage III colon cancer disease-free survival in patients treated by 5-fluorouracil and leucovorin with or without oxaliplatin. Ann Oncol. 2010;21:772–80.
Melling N, Kowitz CM, Simon R, Bokemeyer C, Terracciano L, Sauter G, et al. High Ki67 expression is an independent good prognostic marker in colorectal cancer. J Clin Pathol. 2016;69:209–14.
Auvinen A, Isola J, Visakorpi T, Koivula T, Virtanen S, Hakama M. Overexpression of p53 and long-term survival in colon carcinoma. Br J Cancer. 1994;70:293–6.
Lanza G Jr., Maestri I, Dubini A, Gafa R, Santini A, Ferretti S, et al. p53 expression in colorectal cancer: relation to tumor type, DNA ploidy pattern and short-term survival. Am J Clin Pathol. 1996;105:604–12.
Manne U, Myers RB, Moron C, Poczatek RB, Dillard S, Weiss H, et al. Prognostic significance of Bcl-2 expression and p53 nuclear accumulation in colorectal adenocarcinoma. Int J Cancer. 1997;74:346–58.
Manne U, Weiss HL, Myers RB, Danner OK, Moron C, Srivastava S, et al. Nuclear accumulation of p53 in colorectal adenocarcinoma: prognostic importance differs with race and location of the tumor. Cancer. 1998;83:2456–67.
Ropponen KM, Kellokoski JK, Lipponen PK, Pietilainen T, Eskelinen MJ, Alhava EM, et al. p22/WAF1 expression in human colorectal carcinoma: association withp53, transcription factor AP-2 and prognosis. Br J Cancer. 1999;81:133–40.
Gallego MG, Acenero MJ, Ortega S, Delgado AA, Cantero JL. Prognostic influence of p53 nuclear overexpression in colorectal carcinoma. Dis Colon Rectum. 2000;43:971–5.
Allegra CJ, Paik S, Colangelo LH, Parr AL, Kirsch I, Kim G, et al. Prognostic value of thymidylate synthase, Ki-67, and p53 in patients with Dukes’ B and C colon cancer: a National Cancer Institute-National Surgical Adjuvant Breast and Bowel Project collaborative study. J Clin Oncol. 2003;21:241–50.
Garrity MM, Burgart LJ, Mahoney MR, Windschitl HE, Salim M, Wiesenfeld M, et al. Prognostic value of proliferation, apoptosis, defective DNA mismatch repair, and p53 overexpression in patients with resected Dukes’ B2 or C colon cancer: a North Central Cancer Treatment Group Study. J Clin Oncol. 2004;22:1572–82.
Watson NF, Madjd Z, Scrimegour D, Spendlove I, Ellis IO, Scholefield JH, et al. Evidence that the p53 negative / Bcl-2 positive phenotype is an independent indicator of good prognosis in colorectal cancer: a tissue microarray study of 460 patients. World J Surg Oncol. 2005;3:47.
Wang C, Wang J, Liu H, Fu Z. Tumor suppressor DLC-1 induces apoptosis and inhibits the growth and invasion of colon cancer cells through the Wnt/beta-catenin signaling pathway. Oncol Rep. 2014;31:2270–8.
Kressner U, Lindmark G, Gerdin B, Pahlman L, Glimelius B. Immunohistological p53 staining is of limited value in the staging and prognostic prediction of colorectal cancer. Anticancer Res. 1996;16:951–7.
Nehls O, Klump B, Holzmann K, Lammering G, Borchard F, Gruenagel HH, et al. Influence of p53 status on prognosis in preoperatively irradiated rectal carcinoma. Cancer. 1999;85:2541–8.
Rosati G, Chiacchio R, Reggiardo G, De Sanctis D, Manzione L. Thymidylate synthase expression, p53, bcl-2, Ki-67 and p27 in colorectal cancer: relationships with tumor recurrence and survival. Tumour Biol. 2004;25:258–63.
Katkoori VR, Suarez-Cuervo C, Shanmugam C, Jhala NC, Callens T, Messiaen L, et al. Bax expression is a candidate prognostic and predictive marker of colorectal cancer. J Gastrointest Oncol. 2010;1:76–89.
Kwon HC, Kim SH, Oh SY, Lee S, Kwon KA, Choi HJ, et al. Clinicopathological significance ofp53, hypoxia-inducible factor 1alpha, and vascular endothelial growth factor expression in colorectal cancer. Anticancer Res. 2010;30:4163–8.
Huh JW, Kim HC, Kim SH, Park YA, Cho YB, Yun SH, et al. Mismatch repair system and p53 expression in patients with T1 and T2 colorectal cancer: predictive role of lymph node metastasis and survival. J Surg Oncol. 2014;109:848–52.
Noda M, Okayama H, Kofunato Y, Chida S, Saito K, Tada T, et al. Prognostic role of FUT8 expression in relation to p53 status in stage II and III colorectal cancer. PLoS ONE. 2018;13:e0200315.
Adrover E, Maestro ML, Sanz-Casla MT, del Barco V, Cerdan J, Fernandez C, et al. Expression of high p53 levels in colorectal cancer: a favourable prognostic factor. Br J Cancer. 1999;81:122–6.
Lan YT, Chang SC, Li AF, Lin TC, Chen WS, Jiang JK, et al. p53 protein accumulation as a prognostic marker in sporadic colorectal cancer. Int J Colorectal Dis. 2007;22:499–506.
Noske A, Lipka S, Budczies J, Muller K, Loddenkemper C, Buhr HJ, et al. Combination of p53 expression and p21 loss has an independent prognostic impact on sporadic colorectal cancer. Oncol Rep. 2009;22:3–9.
Munro AJ, Lain S, Lane DP. P53 abnormalities and outcomes in colorectal cancer: a systematic review. Br J Cancer. 2005;92:434–44.
Ricciardiello L, Ceccarelli C, Angiolini G, Pariali M, Chieco P, Paterini P, et al. High thymidylate synthase expression in colorectal cancer with microsatellite instability: implications for chemotherapeutic strategies. Clin Cancer Res. 2005;11:4234–40.
Ogino S, Kawasaki T, Kirkner GJ, Ogawa A, Dorfman I, Loda M, et al. Down-regulation ofp21 (CDKN1A/CIP1) is inversely associated with microsatellite instability and CpG island methylator phenotype (CIMP) in colorectal cancer. J Pathol. 2006;210:147–54.
Nehls O, Okech T, Hsieh CJ, Enzinger T, Sarbia M, Borchard F, et al. Studies onp53, BAX and Bcl-2 protein expression and microsatellite instability in stage III (UICC) colon cancer treated by adjuvant chemotherapy: major prognostic impact of proapoptotic BAX. Br J Cancer. 2007;96:1409–18.
Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–7.
Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–57.
Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–26.
Des Guetz G, Schischmanoff O, Nicolas P, Perret GY, Morere JF, Uzzan B. Does microsatellite instability predict the efficacy of adjuvant chemotherapy in colorectal cancer? A systematic review with meta-analysis. Eur J Cancer. 2009;45:1890–6.
Webber EM, Kauffman TL, O’Connor E, Goddard KA. Systematic review of the predictive effect of MSI status in colorectal cancer patients undergoing 5FU-based chemotherapy. BMC Cancer. 2015;15:156.
Yang B, Eshleman JR, Berger NA, Markowitz SD. Wild-type p53 protein potentiates cytotoxicity of therapeutic agents in human colon cancer cells. Clin Cancer Res. 1996;2:1649–57.
Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, et al. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999;104:263–9.
Longley DB, Allen WL, McDermott U, Wilson TR, Latif T, Boyer J, et al. The roles of thymidylate synthase and p53 in regulating Fas-mediated apoptosis in response to antimetabolites. Clin Cancer Res. 2004;10:3562–71.
Boyer J, McLean EG, Aroori S, Wilson P, McCulla A, Carey PD, et al. Characterization of p53 wild-type and null isogenic colorectal cancer cell lines resistant to 5-fluorouracil, oxaliplatin, and irinotecan. Clin Cancer Res. 2004;10:2158–67.
McDermott U, Longley DB, Galligan L, Allen W, Wilson T, Johnston PG. Effect of p53 status and STAT1 on chemotherapy-induced, Fas-mediated apoptosis in colorectal cancer. Cancer Res. 2005;65:8951–60.
Toscano F, Parmentier B, Fajoui ZE, Estornes Y, Chayvialle JA, Saurin JC, et al. p53 dependent and independent sensitivity to oxaliplatin of colon cancer cells. Biochem Pharmacol. 2007;74:392–406.
Benhattar J, Cerottini JP, Saraga E, Metthez G, Givel JC. p53 mutations as a possible predictor of response to chemotherapy in metastatic colorectal carcinomas. Int J Cancer. 1996;69:190–2.
Soong R, Powell B, Elsaleh H, Gnanasampanthan G, Smith DR, Goh HS, et al. Prognostic significance of TP53 gene mutation in 995 cases of colorectal carcinoma. Influence of tumour site, stage, adjuvant chemotherapy and type of mutation. Eur J Cancer. 2000;36:2053–60.
Allegra CJ, Parr AL, Wold LE, Mahoney MR, Sargent DJ, Johnston P, et al. Investigation of the prognostic and predictive value of thymidylate synthase, p53, and Ki-67 in patients with locally advanced colon cancer. J Clin Oncol. 2002;20:1735–43.
Elsaleh H, Powell B, Soontrapornchai P, Joseph D, Goria F, Spry N, et al. p53 gene mutation, microsatellite instability and adjuvant chemotherapy: impact on survival of 388 patients with Dukes’ C colon carcinoma. Oncology. 2000;58:52–9.
Popat S, Chen Z, Zhao D, Pan H, Hearle N, Chandler I, et al. A prospective, blinded analysis of thymidylate synthase and p53 expression as prognostic markers in the adjuvant treatment of colorectal cancer. Ann Oncol. 2006;17:1810–7.
Russo A, Bazan V, Iacopetta B, Kerr D, Soussi T, Gebbia N, et al. The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: influence of tumor site, type of mutation, and adjuvant treatment. J Clin Oncol. 2005;23:7518–28.
Allred DC, Clark GM, Elledge R, Fuqua SA, Brown RW, Chamness GC, et al. Association of p53 protein expression with tumor cell proliferation rate and clinical outcome in node-negative breast cancer. J Natl Cancer Inst. 1993;85:200–6.
Mouradov D, Sloggett C, Jorissen RN, Love CG, Li S, Burgess AW, et al. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 2014;74:3238–47.
Domingo E, Camps C, Kaisaki PJ, Parsons MJ, Mouradov D, Pentony MM, et al. Mutation burden and other molecular markers of prognosis in colorectal cancer treated with curative intent: results from the QUASAR 2 clinical trial and an Australian community-based series. Lancet Gastroenterol Hepatol. 2018;3:635–43.
Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–57.
Lindor NM, Burgart LJ, Leontovich O, Goldberg RM, Cunningham JM, Sargent DJ, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002;20:1043–8.
Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–67.
Lowe SW, Bodis S, McClatchey A, Remington L, Ruley HE, Fisher DE, et al. p53 status and the efficacy of cancer therapy in vivo. Science. 1994;266:807–10.
Friedlander P, Haupt Y, Prives C, Oren M. A mutant p53 that discriminates between p53-responsive genes cannot induce apoptosis. Mol Cell Biol. 1996;16:4961–71.
Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–30.
Klump B, Nehls O, Okech T, Hsieh CJ, Gaco V, Gittinger FS, et al. Molecular lesions in colorectal cancer: impact on prognosis? Original data and review of the literature. Int J Colorectal Dis. 2004;19:23–42.
Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–34.
Acknowledgements
The authors thank the Victorian Cancer BioBank and Biogrid Australia for provision of patient specimens and clinical data. This work was supported by grant #1120882, awarded through the Priority-driven Collaborative Cancer Research Scheme and funded by Cancer Australia (to O.M.S), the Victorian Government’ Operational Infrastructure Support Program, a University of Melbourne Department of Pathology Career Development Award (to D.S.W.) and an Austin Medical Research Foundation Project Grant (to D.S.W). OMS (GNT1136119) and JMM (GNT104692) are NHMRC Senior Research Fellows.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Williams, D.S., Mouradov, D., Browne, C. et al. Overexpression of TP53 protein is associated with the lack of adjuvant chemotherapy benefit in patients with stage III colorectal cancer. Mod Pathol 33, 483–495 (2020). https://doi.org/10.1038/s41379-019-0353-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-019-0353-2
This article is cited by
-
Potentials and future perspectives of multi-target drugs in cancer treatment: the next generation anti-cancer agents
Cell Communication and Signaling (2024)