Abstract
Short-read next generation sequencing (NGS) has become the predominant first-line technique used to diagnose patients with rare genetic conditions. Inherent limitations of short-read technology, notably for the detection and characterization of complex insertion-containing variants, are offset by the ability to concurrently screen many disease genes. “Third-generation” long-read sequencers are increasingly being deployed as an orthogonal adjunct technology, but their full potential for molecular genetic diagnosis has yet to be exploited. Here, we describe three diagnostic cases in which pathogenic mobile element insertions were refractory to characterization by short-read sequencing. To validate the accuracy of the long-read technology, we first used Sanger sequencing to confirm the integration sites and derive curated benchmark sequences of the variant-containing alleles. Long-read nanopore sequencing was then performed on locus-specific amplicons. Pairwise comparison between these data and the previously determined benchmark alleles revealed 100% identity of the variant-containing sequences. We demonstrate a number of technical advantages over existing wet-laboratory approaches, including in silico size selection of a mixed pool of amplification products, and the relative ease with which an automated informatics workflow can be established. Our findings add to a growing body of literature describing the diagnostic utility of long-read sequencing.
Similar content being viewed by others
Introduction
The universal adoption of short-read next generation sequencers by both research and diagnostic laboratories has transformed the availability and repertoire of molecular genetic assays. The ability to sequence multiple target loci concurrently, rather than consecutively, has increased diagnostic throughput and helped refine the mutation spectra of hundreds of genetic disorders. The overwhelming majority of sequences are generated using sequencing-by-synthesis chemistry on instruments manufactured by Illumina, Inc. However, methods for the pre-sequencing enrichment of target DNA sequences vary widely, with the chosen approach typically being dependent on the scope of the test. (This can range from small panels of specified genes through to the analysis of whole exomes or genomes.) No matter which enrichment methods are used, a trade-off exists between the amount of sequence targeted for analysis (an assay’s “genomic footprint”) and the test sensitivity (which depends in some fashion on the cumulative read depth at a given locus).
Increasingly specialized tools have been developed to address some of the nuances of short-read datasets, enabling, for example, the characterization of repeat expansions and mobile element insertions [1, 2], albeit with varying success. Although such tools improve variant detection capabilities, the need to undertake extensive validation studies, before the analysis can be considered robust or accredited, means that their rapid incorporation into existing diagnostic data processing pipelines may not be possible. Consequently, although we and others have previously demonstrated how reprocessing of legacy diagnostic datasets, using updated software tools can (unsurprisingly) lead to improved diagnostic yield [3], efforts to retrospectively investigate routine diagnostic referrals remain limited, and are still typically only undertaken at large, centrally coordinated, genomics centers.
“Third generation” long-read single molecule sequencing offers a way to overcome some of the inherent limitations and technical challenges of short-read sequencing, enabling analysis of so-called “dark” or “camouflaged” genomic regions [4] and thereby improving the quality and sensitivity of genomic assays. One well-recognized drawback of short-read sequencing is the inability to obtain unambiguous alignments to the reference sequence when analyzing some genomic regions. This can, for example, prevent differentiation between gene- and pseudogene-derived sequences. A diagnostic consequence of this scenario is that pathogenic gene conversion events can be masked by preferential alignment of short-reads to the pseudogene locus, as we recently reported at the TMEM231 locus in Meckel–Gruber syndrome (OMIM:249000) [5]. Another frequent area of difficulty concerns insertion variants; the presence of low complexity repeat sequences at the integration site and the generation of two sets of read-pairs (spanning the variant’s 5′ and 3′ junction sequences), are both factors that can create interpretative challenges. Regardless of the variant detection algorithm used, when the number of inserted nucleotides exceeds the sequencer’s maximum read length, characterization of the intervening sequence will remain incomplete.
Additional analytical constraints are imposed by the short-read sequencing chemistry itself; regions of high GC content (characteristically including a gene’s first exon) are often associated with poor amplification and a reduced cumulative read depth. Long-read sequencing protocols, using PCR-free approaches, have addressed this challenge, although the requirement for a large mass of input DNA, and the relative inefficiency of target enrichment reactions has, to-date, limited the widespread adoption of these methods [6]. Nevertheless, for tandem-repeat disorders targeted analysis of native DNA strands presents the exciting prospect that both the size and methylation status of an expanded allele can be determined in a single assay [7].
Here, we present an assessment of the utility of “third-generation” nanopore sequencing for the confirmation and characterization of mobile element insertions. For three cases in which pathogenic insertion-containing variants were identified, we designed locus-specific PCR amplicons based on the integration sites identified in the short-read sequencing dataset. Sanger sequencing of amplification products from patient DNA then enabled determination of a benchmark sequence of the insertion-containing allele. De novo assembly of long reads generated by nanopore sequencing was performed and the accuracy of the consensus sequence was assessed following pairwise alignment with the benchmark sequence. We discuss how implementation of long-read nanopore sequencing can offer benefits over existing molecular approaches.
Materials and methods
The cases had been referred to the Yorkshire Regional Genetics Laboratory for diagnostic analysis of hereditary neoplasia. DNA was isolated from peripheral blood lymphocytes using a bead-based extraction method, following written consent. Ethical approval for this study was granted by the Leeds East Research Ethics Committee (18/YH/0070).
Short-read next-generation sequencing
Short-read sequencing was used to screen the coding sequences and invariant splice sites of target genes specified by the referring clinician. For cases 1 and 2, a custom SureSelect hybridization reagent was used to perform enrichment of 155 target genes (Agilent Technologies, Wokingham, UK). Illumina-compatible sequencing libraries were created following manufacturer’s protocols as previously described [3]. Sixteen libraries were pooled in equimolar amounts to create a library pool which was combined with additional library pools and sequenced on a NextSeq500 instrument (Illumina, San Diego, CA). Paired-end 151-bp sequence reads were generated using a high-output reagent cartridge. Raw data conversion and read demultiplexing were performed using bcl2fastq v.2.17.1.14 (Illumina).
For case 3, target enrichment of BRCA1 and BRCA2 was performed using a previously described LR-PCR workflow [8]. Forty libraries were sequenced on a MiSeq, generating 151-bp paired end reads. Sequencing and raw data conversion, from bcl to FASTQ.gz format, were performed using Real-Time Analysis v.1.18.42 and MiSeq Control Software v.2.3.0.3 (Illumina). Demultiplexing of library-specific reads defined using 6-bp barcodes located at the 3′ ends of adapter molecules was performed with fastq-multx v.1.02.772 (https://expressionanalysis.github.io/ea-utils/).
Sequence reads from all three cases were processed using an in-house pipeline. Adapter sequences and low-quality bases (-q 10) were first trimmed from each read-pair using Cutadapt v.2.4 (https://cutadapt.readthedocs.org) [9] before being aligned to an indexed human reference genome (build hg19) using BWA MEM v.0.7.17 (http://bio-bwa.sourceforge.net) [10]. Picard v.2.20.5-0 (http://broadinstitute.github.io/picard) was used to perform sam-to-bam conversion, duplicate marking and bam file indexing. Assembly-based realignment was performed over each target interval using ABRA2 v.0.09 (http://github.com/mozack/abra2) [11]. The Genome Analysis Toolkit (GATK) v.3.7-0 was used to perform indel realignment, base quality score recalibration and variant calling using the HaplotypeCaller, following the conventions described in the GATK best practice workflow (https://software.broadinstitute.org/gatk/) [12]. Identified variants were annotated with functional information and allele frequency data using Alamut Batch standalone v.1.11 (database version 2020.03.18; Interactive Biosoftware, Rouen, France). For cases 1 and 2, copy number variants were assessed using a comparative read-depth approach, with normalized read counts from the control group being obtained from intra-batch patient libraries. The resulting dataset was interpreted following Association for Clinical Genomic Science best practice guidelines [13]. To assess assay performance, coverage metrics were generated using the GATK walkers DepthOfCoverage, CallableLoci and CountReads. Aligned sequence reads were examined using the Integrative Genome Viewer v.2.4.10 (http://software.broadinstitute.org/software/igv/) [14] and soft-clipped reads were interrogated using BLAT (http://genome.ucsc.edu/cgi-bin/hgBlat) [15]. To enable detection of mobile element insertions MELT v.2.2.0 was retrospectively implemented (https://melt.igs.umaryland.edu/index.php) [2].
Dye-terminator capillary sequencing
To confirm the precise sequence of each Alu insertion, PCR amplicons were first optimized. Each reaction consisted of 1 μL of genomic DNA (~50 ng/μL), 10.6 μL MegaMix PCR reagents (Microzone Ltd., Haywards Heath, UK) and 0.2 μL each of 10 μM locus specific forward and reverse primers (Thermo Fisher Scientific, Waltham, MA) as detailed in Supplementary Table S1. The standardized thermocycling conditions recorded in Supplementary Table S2 were applied to all reactions. Amplification products were resolved on a 2% Tris-borate-EDTA agarose gel, before being extracted and purified using a QIAquick column (Qiagen GmbH, Hilden, Germany). All primer sets contained universal sequencing tags which enabled Sanger sequencing on an ABI3730 using our standard laboratory workflow. Manufacturer’s protocols were followed throughout (Life Technologies Ltd., Paisley, UK). Sequence chromatograms were analyzed using 4Peaks software v.1.8 (http://nucleobytes.com/4peaks/index.html).
Long-read sequencing
Nanopore sequencing was performed on amplification products purified by AMPure XP bead clean-up (Beckman Coulter, Bucks, UK). A separate sequencing library was created for each case. An end-repair and nickase treatment reaction was first performed, which consisted of 1.75 μL UltraTM II end prep reaction buffer (New England Biolabs [NEB], Ipswich, MA), 1.75 μL FFPE DNA repair buffer (NEB), 1.5 μL UltraTM II end prep enzyme mix (NEB), 1.0 μL FFPE DNA repair mix (NEB), 100 fmol PCR product and nuclease-free water to make a total volume of 24 μL. The reaction was incubated at 20 °C for 5 min and then 65 °C for 5 min. An AMPure XP bead clean-up (Beckman Coulter) was performed before sequencing adapters were ligated to the double-stranded DNA. The reaction comprised 30 μL of PCR products, 12.5 μL of Ligation Buffer (Oxford Nanopore Technologies [ONT], Oxford, UK), 5.0 μL of Quick Ligase (NEB) and 2.5 μL of Adapter Mix (ONT). The reaction was incubated at room temperature for 10 min. A further AMPure XP bead clean-up was performed; Short Fragment Buffer (ONT) was used to wash the beads before the sample was eluted in 15 μL of Elution Buffer (ONT). A Flongle flowcell was prepared for sequencing using 120 μL of flowcell priming mix (3 μL of Flush Tether (ONT) was combined with 117 μL of Flush Buffer (ONT) which was loaded through the priming port. Twenty fmol of the library was combined with 15 μL of Sequencing Buffer (ONT) and 10 μL of Loading Beads (ONT) then loaded into the flowcell. A 24-h Flongle sequencing run was initiated using MinKNOW software (v.3.6.5; ONT).
Offline basecalling was performed using Guppy (v.3.6.0) to convert raw data from fast5 to FASTQ format (http://nanoporetech.com). Adapter sequences were trimmed from the resulting reads using Porechop (v.0.2.3) (https://github.com/rrwick/Porechop) before NanoFilt (v.2.2.0) removed low quality reads and performed selection based on read length (https://github.com/wdecoster/nanofilt) [16]. NanoStat (v.1.1.2) was used to calculate summary read metrics (https://github.com/wdecoster/nanostat). A consensus de novo assembly was generated using Canu (v.2.0) [17]. Pairwise sequence alignment between curated variant-containing benchmark alleles and de novo consensus assemblies was performed using the Needleman-Wunsch algorithm (https://www.ebi.ac.uk/Tools/psa/emboss_needle/) [18].
Results
The three cases described were all referred to the Yorkshire and North East Genomic Laboratory Hub for mutation screening of hereditary cancer genes. Case 1 presented with a neck paraganglioma and was found to have raised plasma 3-methoxytyramine (810 pM, reference range: 0–185 pM). She was referred for analysis of pheochromocytoma/paraganglioma-predisposing genes (EGLN1, EGLN2, KIF1B, MAX, NF1, RET, SDHA, SDHAF2, SDHB, SDHC, SDHD, TMEM127, and VHL). Case 2 presented with a clinical diagnosis of neurofibromatosis type 1 and was referred for genetic analysis of café-au-lait macule-associated disorder genes (BAP1, LZTR1, NF1, NF2, PTEN, SMARCB1, SMARCE1, SPRED1, and SUFU). Case 3 was referred for mutation screening of hereditary breast cancer genes BRCA1 and BRCA2. Either custom hybridization (Cases 1 and 2) or long-range PCR target enrichment (Case 3), followed by short read next generation sequencing, was performed. Assay performance metrics for the three sequencing libraries are recorded in Supplementary Table S3. Initial analysis of these datasets was negative in all three cases, no disease-associated variants having been identified using the laboratory’s standard variant-calling workflow.
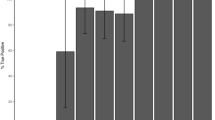
Second-line analysis was directed at the detection of possible copy number changes in the target loci. For Case 1, comparative read depth analysis revealed a reduced dosage quotient (0.67) in SDHB exon 3 (NM_003000.3) suggestive of a heterozygous deletion. Additional analysis of Case 2, by multiplex ligation-dependent probe amplification (MLPA) of NF1 revealed an apparent single-exon deletion of exon 53 (NM_000267.3) (the comparative read depth analysis dosage quotient for this exon was 0.81). However, manual scrutiny of the aligned sequence reads, at the putative deletion-containing loci (Cases 1 and 2), and for Case 3, the BRCA2 exon 3 (NM_000059.4) Portuguese founder mutation locus, revealed the presence of mobile element insertions intersecting the coding sequence. The appearances are shown in Fig. 1; in each case the variant shows up as a short region of increased per-base read depth (due to micro-duplications at the integration site) located between two distinct sets of flanking soft-clipped reads; one set defining the variant-specific sequences and the other set running into the terminal poly(A) tail.
a SDHB exon 3 b NF1 exon 53, and c BRCA2 exon 3. Reads are collapsed and “viewed as pairs” using the Integrative Genomics Viewer with “quick-consensus mode” enabled. Mismatched bases, representing soft-clipped alignments, are colored with respect to the sense strand (T, red; C, blue; A, Green; and G, brown). The “plateau” of increased read coverage coincides approximately with the duplicated nucleotides at the integration site. The y-axis scale for each cumulative read depth plot is labeled. Arrows denote the direction of transcription. Coding nucleotide numbering corresponds to the reported transcript for each gene.
To estimate the number of inserted nucleotides, locus-specific assays were devised and PCR amplification products were resolved using an automated electrophoresis system (Supplementary Fig. S1). We next determined the integration sites and full-length sequences of each inserted mobile element, by Sanger sequencing the variant-containing alleles. The detailed structures of each allele, assembled from forward and reverse chromatograms, are displayed in Fig. 2. Each inserted sequence was 100% identical to a known Alu family member. All three mobile element insertions intersected the coding sequence of the target gene, and 11–12 bp sequence duplications were identified at each variant’s integration site. Although the mobile elements inserted in cases 1 and 2 are both of the AluYb8 subtype, pairwise comparison of their sequences revealed them to be non-identical (285/287 matching bases).
a SDHB exon 3 locus b NF1 exon 53 locus, and c BRCA2 exon 3 locus. Exon sequences are shaded blue, intron sequences remain unshaded and the mobile element insertions are shaded green. Genomic coordinates are reported according to human reference genome build hg19. Coding nucleotides are reported according to the following transcripts: SDHB, NM_003000.3; NF1, NM_000267.3; and BRCA2, NM_000059.4.
The integration site of the BRCA2 variant c.156_157insAlu, and its classification as an AluYa5 subtype, are consistent with previous reports, suggesting that case 3 shares ancestry with the reported Portuguese founder population [19].
To assess the utility of long-read sequencing for simple determination of the structure of the insertion alleles, we carried out nanopore sequencing on Flongle flowcells. Ligation-based library preparation was performed on amplification products generated by locus-specific PCR reactions. Summary run metrics are recorded in Table 1. Rather than having to physically purify the two alleles (as for Sanger sequencing), adapter-trimmed reads were simply filtered by length to select those from the variant-containing allele and to exclude PCR concatamers. We performed de novo assembly of the quality-filtered reads and then pairwise alignment to the Sanger-sequencing-verified benchmark nucleotide sequence of the locus. For each locus, the alignments were highly concordant (>99%) see Supplementary Fig. S2; the individual values were 100% (SDHB locus), 99.5% (NF1 locus), and 99.1% (BRCA2 locus). Observed mismatches were either due to absent terminal nucleotides (possibly caused by stringent adapter trimming), or at a 10-bp poly(T) tract in the BRCA2 locus, consequent upon an underestimate of T nucleotides in the consensus assembly. To appraise the value of individual raw nanopore reads, we extracted the read with the highest mean basecall quality score (Q) and length (L), from each locus-specific dataset. Pairwise alignment between these individual reads and the curated benchmark sequences yielded identity scores of 95.7% (SDHB locus, Q:14, L: 591 bp), 95.1% (NF1 locus, Q:21, L: 599 bp), and 95.4% (BRCA2 locus, Q:16, L: 752 bp) respectively.
Discussion
Short-read, highly parallel NGS workflows have become the de facto method of choice for identifying pathogenic, disease-causing variants in a diagnostic setting. For genetically heterogeneous conditions, there has been much debate about which genes should be included in diagnostic “panels”, as well as the relative merits of focusing on narrow or broad clinical referral criteria. International working groups comprising expert panel members have been established across all clinical domains through the ClinGen initiative [20] and tools such as Panel App aim to broker community consensus and a standardization of approach [21]. An emerging theme is that whole genome sequencing, implemented cheaply at large scale, has the potential to outdate discussions concerning the configuration of physical enrichment reagents (custom panels versus exomes), and major strategic financial investments in support of this philosophy have been made [22]. However, this “one size fits all” strategy is being undermined by growing evidence highlighting the importance of “third-generation” sequencing, particularly for the identification of complex structural variants [23].
To capitalize on the massive throughput of short-read NGS (allowing parallel testing of multiple samples across large genomic footprints), automated variant detection algorithms have had to be deployed. While this largely eliminates the onerous task of manually inspecting raw sequence reads for the presence of non-reference bases, laboratories must instead rely on their informatics pipelines for variant detection; the sensitivity of these pipelines depends on the mutation spectrum for which they have been tailored. We have described here three cases in which mobile element insertions were (unsurprisingly) refractory to detection using a standard diagnostic pipeline optimized for the detection of single nucleotide variants and small indels (insertion-deletion variants). In each case, manual inspection of short-read NGS data was required for clues to the true nature of the pathogenic alleles. For Case 1, the manual inspection was prompted by a relative reduction in the number of NGS reads aligned to SDHB exon 3. This reduced read coverage resulted not from a deletion, but from impaired alignment of insertion-containing reads. Whether such events are detectable by comparative dosage analysis is likely to depend on the size of the defined target exon. (A proportionally smaller reduction in coverage will be evident for larger exons.) This sensitivity problem is exemplified by Case 2, for which the dosage quotient (0.81) was at the low end of the normal range (0.8–1.2) and the pathogenic NF1 exon 53 insertion was only identified due to the serendipitous disruption of an MLPA probe-binding site.
The iterative improvement of NGS data processing pipelines depends on the detailed understanding of edge case requirements; here, for example, we were able to retrospectively calibrate MELT for identification of the reported variants. We note that this application’s rapid runtime makes it ideally suited for retrospective analysis of legacy short-read datasets. Such work is however predicated on the availability of suitable test cases. Resources such as the reference materials and benchmarking datasets developed by the Genome in a Bottle consortium [24] are of great importance in this regard, but the need continues for further shared datasets in which atypical classes of sequence variation are represented.
Previous efforts to identify mobile element insertions in large clinical cohorts have indicated that their contribution to disease is low though significant (~0.04% in severe developmental disorders) [25]. We speculate that the difficulties associated with detecting and characterizing mobile element insertions mean that they are likely to be underrepresented in both population and disease-specific databases. Because Sanger sequencing requires physical separation of alleles and manual inspection of chromatograms to verify and curate individual sequences, it is a laborious and error prone task. Our analysis of the long-read dataset utilized Canu to perform a de novo consensus assembly across target loci. The informatics pipeline is highly automated, requires little user intervention and is readily scalable. While de novo assembly is a technique commonly utilized by the bacteriology and metagenomics communities, it has seldom been applied to human genomic datasets. This highlights the ongoing potential for diagnostic capabilities to be enriched by the introduction of technologies from other disciplines.
As exemplified here, the full characterization of mobile element insertions identified from short-read NGS datasets requires follow-up analyses that may exceed the scope of testing in many diagnostic laboratories. Although Sanger sequencing is typically chosen for such follow-up investigations, terminal poly(A) tracts (which induce polymerase slippage and uninterpretable sequence chromatograms) often preclude sequencing from the 3′ end of the element. It is also often necessary to gel purify the insertion-containing allele to prevent interference from chromatogram peaks in a mixed population of amplification products. In contrast, we have found that nanopore sequencing of a mixed pool of amplification products (comprising both normal and mutated alleles) can be analyzed using an in silico size selection approach to isolate insertion-containing reads.
Few studies of targeted nanopore sequencing and its application to molecular diagnostics have been reported to-date. Our experience demonstrates high concordance between a benchmark Sanger sequence and the nanopore dataset, suggesting that Flongle-based nanopore sequencing could be routinely deployed for the characterization and confirmation of challenging sequence variants. Although the highest quality single-molecule read did not individually match the benchmark sequence perfectly, it was still sufficiently accurate to identify the class of inserted repeat sequence. Despite a 58-fold difference in yield between worst and best performing nanopore runs, we saw no reduction in consensus accuracy across the insertion sequence. Indeed, the only reduction in pairwise identity was seen at the primer binding sites (likely attributable to stringent adapter trimming) and for the BRCA2 amplicon at a 10-bp poly(T) tract. The considerable number of reads available following length and quality filtering suggests that the workflow could be further adapted to multiplexing individual patient libraries.
In summary, we report how retrospective analysis of NGS datasets can be used to calibrate specific informatics tools. We further demonstrate that low-cost nanopore based sequencing is able to determine full-length Alu-repeat insertions, with a simplified workflow and a consensus accuracy that is comparable to Sanger sequencing, the existing gold-standard approach.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Dashnow H, Lek M, Phipson B, Halman A, Sadedin S, Lonsdale A, et al. STRetch: detecting and discovering pathogenic short tandem repeat expansions. Genome Biol. 2018;19:121.
Gardner EJ, Lam VK, Harris DN, Chuang NT, Scott EC, Pittard WS, et al. The Mobile Element Locator Tool (MELT): population-scale mobile element discovery and biology. Genome Res. 2017;27:1916–29.
Watson CM, Camm N, Crinnion LA, Clokie S, Robinson RL, Adlard J, et al. Increased sensitivity of diagnostic mutation detection by re-analysis incorporating local reassembly of sequence reads. Mol Diagn Ther. 2017;21:685–92.
Ebbert MTW, Jensen TD, Jansen-West K, Sens JP, Reddy JS, Ridge PG, et al. Systematic analysis of dark and camouflaged genes reveals disease-relevant genes hiding in plain sight. Genome Biol. 2019;20:97.
Watson CM, Dean P, Camm N, Bates J, Carr IM, Gardiner CA, et al. Long-read nanopore sequencing resolves a TMEM231 gene conversion event causing Meckel-Gruber syndrome. Hum Mutat. 2020;41:525–31.
Watson CM, Crinnion LA, Hewitt S, Bates J, Robinson R, Carr IM, et al. Cas9-based enrichment and single-molecule sequencing for precise characterization of genomic duplications. Lab Investig. 2020;100:135–46.
Giesselmann P, Brändl B, Raimondeau E, Bowen R, Rohrandt C, Tandon R, et al. Analysis of short tandem repeat expansions and their methylation state with nanopore sequencing. Nat Biotechnol. 2019;37:1478–81.
Morgan JE, Carr IM, Sheridan E, Chu CE, Hayward B, Camm N, et al. Genetic diagnosis of familial breast cancer using clonal sequencing. Hum Mutat. 2010;31:484–91.
Martin M. Cutadapt removes adapter sequences from high‐ throughput sequencing reads. EMBnet J. 2011;17:10–2.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
Mose LE, Perou CM, Parker JS. Improved indel detection in DNA and RNA via realignment with ABRA2. Bioinformatics. 2019;35:2966–73.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8.
Ellard S, Baple EL, Callaway A, Berry I, Forrester N, Turnbull C, et al. ACGS best practice guidelines for variant classification in rare disease (2020). Association for Clinical Genomic Science. https://www.acgs.uk.com/media/11631/uk-practice-guidelines-for-variant-classification-v4-01-2020.pdf.
Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92.
Kent WJ. BLAT-the BLAST-like Alignment Tool. Genome Res. 2002;12:656–64.
De Coster W, D’Hert S, Schultz DT, Cruts M, Van Broeckhoven C. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics. 2018;34:2666–9.
Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–36.
Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–41.
Machado PM, Brandão RD, Cavaco BM, Eugénio J, Bento S, Nave M, et al. Screening for a BRCA2 rearrangement in high-risk breast/ovarian cancer families: evidence for a founder effect and analysis of the associated phenotypes. J Clin Oncol. 2007;25:2027–34.
Rehm HL, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, et al. ClinGen-the clinical genome resource. N Engl J Med. 2015;372:2235–42.
Martin AR, Williams E, Foulger RE, Leigh S, Daugherty LC, Niblock O, et al. PanelApp crowdsources expert knowledge to establish consensus diagnostic gene panels. Nat Genet. 2019;51:1560–5.
Turnbull C, Scott RH, Thomas E, Jones L, Murugaesu N, Pretty FB, et al. The 100,000 Genomes Project: bringing whole genome sequencing to the NHS. BMJ. 2018;361:k1687.
Logsdon GA, Vollger MR, Eichler EE. Long-read human genome sequencing and its applications. Nat Rev Genet. 2020;21:597–614.
Zook JM, McDaniel J, Olson ND, Wagner J, Parikh H, Heaton H, et al. An open resource for accurately benchmarking small variant and reference calls. Nat Biotechnol. 2019;37:561–6.
Gardner EJ, Prigmore E, Gallone G, Danecek P, Samocha KE, Handsaker J, et al. Contribution of retrotransposition to developmental disorders. Nat Commun. 2019;10:4630.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Dr Watson has received travel expenses to speak at an Oxford Nanopore Technologies organized conference.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Watson, C.M., Crinnion, L.A., Lindsay, H. et al. Assessing the utility of long-read nanopore sequencing for rapid and efficient characterization of mobile element insertions. Lab Invest 101, 442–449 (2021). https://doi.org/10.1038/s41374-020-00489-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-020-00489-y
This article is cited by
-
Applications of long-read sequencing to Mendelian genetics
Genome Medicine (2023)
-
Mastering DNA chromatogram analysis in Sanger sequencing for reliable clinical analysis
Journal of Genetic Engineering and Biotechnology (2023)
-
Long-Read Nanopore Sequencing of RPGR ORF15 is Enhanced Following DNase I Treatment of MinION Flow Cells
Molecular Diagnosis & Therapy (2023)