Abstract
Objective
We describe our experience with whole-exome sequencing (WES) in fetuses with central nervous system (CNS) abnormalities following a normal chromosomal microarray result.
Methods
During the study period (2014–2017) 7 cases (9 fetuses) with prenatally diagnosed CNS abnormality, whose chromosomal microarray analysis was negative, were offered whole-exome sequencing analysis.
Results
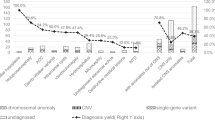
A pathogenic or a likely pathogenic variant was found in 5 cases including a previously described, likely pathogenic de novo TUBA1A variant (Case #1); a previously described homozygous VRK1 variant (Case #2); an X-linked ARX variant (Case #3); a likely pathogenic heterozygous variant in the TUBB3 gene (Case #5). Finally, in two fetuses of the same couple (Case #6), a compound heterozygous state was detected, consisting of the NPHP1 gene deletion and a sequence variant of uncertain significance. Two additional cases had normal WES results.
Conclusion
Whole-exome sequencing may improve prenatal diagnosis in fetuses with CNS abnormalities.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Onkar D, Onkar P, Mitra K. Evaluation of fetal central nervous system anomalies by ultrasound and its anatomical co-relation. J Clin Diagn Res. 2014;8:AC05–07.
Sun L, Wu Q, Jiang SW, Yan Y, Wang X, Zhang J, et al. Prenatal diagnosis of central nervous system anomalies by high-resolution chromosomal microarray analysis. Biomed Res Int. 2015;2015:426379.
Huang J, Wah IY, Pooh RK, Choy KW. Molecular genetics in fetal neurology. Semin Fetal Neonatal Med. 2012;17:341–6.
Drury S, Williams H, Trump N, Boustred C, Lench N, Scott RH, et al. Exome sequencing for prenatal diagnosis of fetuses with sonographic abnormalities. Prenat Diagn. 2015;35:1010–7.
Beaudet AL. Using fetal cells for prenatal diagnosis: history and recent progress. Am J Med Genet C Semin Med Genet. 2016;172:123–7.
Du L, Xie HN, Huang LH, Xie YJ, Wu LH. Prenatal diagnosis of submicroscopic chromosomal aberrations in fetuses with ventricular septal defects by chromosomal microarray-based analysis. Prenat Diagn. 2016;36:1178–84.
Yang X, Li R, Fu F, Zhang Y, Li D, Liao C. Submicroscopic chromosomal abnormalities in fetuses with increased nuchal translucency and normal karyotype. J Matern Fetal Neonatal Med. 2017;30:194–8.
Best S, Wou K, Vora N, Van der Veyver IB, Wapner R, Chitty LS. Promises, pitfalls and practicalities of prenatal whole exome sequencing. Prenat Diagn. 2017;38:10–19.
de Wit MC, Srebniak MI, Govaerts LC, Van Opstal D, Galjaard RJ, Go AT. Additional value of prenatal genomic array testing in fetuses with isolated structural ultrasound abnormalities and a normal karyotype: a systematic review of the literature. Ultrasound Obstet Gynecol. 2014;43:139–46.
Charan P, Woodrow N, Walker SP, Ganesamoorthy D, McGillivray G, Palma-Dias R. High-resolution microarray in the assessment of fetal anomalies detected by ultrasound. Aust N Z J Obstet Gynaecol. 2014;54:46–52.
Malinger G, Kidron D, Schreiber L, Ben-Sira L, Hoffmann C, Lev D, et al. Prenatal diagnosis of malformations of cortical development by dedicated neurosonography. Ultrasound Obstet Gynecol. 2007;29:178–91.
Manzini MC, Walsh CA. What disorders of cortical development tell us about the cortex: one plus one does not always make two. Curr Opin Genet Dev. 2011;21:333–9.
Saunders CJ, Miller NA, Soden SE, Dinwiddie DL, Noll A, Alnadi NA, et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med. 2012;4:154ra135.
Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369:1502–11.
Vora NL, Powell B, Brandt A, Strande N, Hardisty E, Gilmore K, et al. Prenatal exome sequencing in anomalous fetuses: New opportunities and challenges. Genet Med. 2017;19:1207–16.
Lei TY, Fu F, Li R, Wang D, Wang RY, Jing XY, et al. Whole-exome sequencing for prenatal diagnosis of fetuses with congenital anomalies of the kidney and urinary tract. Nephrol Dial Transplant. 2017;32:1665–75.
Abou Tayoun AN, Spinner NB, Rehm HL, Green RC, Bianchi DW. Prenatal DNA sequencing: clinical, counseling, and diagnostic laboratory considerations. Prenat Diagn. 2017;38:26–32.
Kehrer C, Hoischen A, Menkhaus R, Schwab E, Muller A, Kim S, et al. Whole exome sequencing and array-based molecular karyotyping as aids to prenatal diagnosis in fetuses with suspected simpson-golabi-behmel syndrome. Prenat Diagn. 2016;36:961–5.
Casey J, Flood K, Ennis S, Doyle E, Farrell M, Lynch SA. Intra-familial variability associated with recessive ryr1 mutation diagnosed prenatally by exome sequencing. Prenat Diagn. 2016;36:1020–6.
Ellard S, Kivuva E, Turnpenny P, Stals K, Johnson M, Xie W, et al. An exome sequencing strategy to diagnose lethal autosomal recessive disorders. Eur J Hum Genet. 2015;23:401–4.
Drury S, Boustred C, Tekman M, Stanescu H, Kleta R, Lench N, et al. A novel homozygous ercc5 truncating mutation in a family with prenatal arthrogryposis--further evidence of genotype-phenotype correlation. Am J Med Genet A. 2014;164A:1777–83.
Carss KJ, Hillman SC, Parthiban V, McMullan DJ, Maher ER, Kilby MD, et al. Exome sequencing improves genetic diagnosis of structural fetal abnormalities revealed by ultrasound. Hum Mol Genet. 2014;23:3269–77.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the american college of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17:405–24.
Gonzaga-Jauregui C, Lotze T, Jamal L, Penney S, Campbell IM, Pehlivan D, et al. Mutations in vrk1 associated with complex motor and sensory axonal neuropathy plus microcephaly. JAMA Neurol. 2013;70:1491–8.
Renbaum P, Kellerman E, Jaron R, Geiger D, Segel R, Lee M, et al. Spinal muscular atrophy with pontocerebellar hypoplasia is caused by a mutation in the vrk1 gene. Am J Hum Genet. 2009;85:281–9.
Vinograd-Byk H, Sapir T, Cantarero L, Lazo PA, Zeligson S, Lev D, et al. The spinal muscular atrophy with pontocerebellar hypoplasia gene vrk1 regulates neuronal migration through an amyloid-beta precursor protein-dependent mechanism. J Neurosci. 2015;35:936–42.
Parisi MA, Bennett CL, Eckert ML, Dobyns WB, Gleeson JG, Shaw DW, et al. The nphp1 gene deletion associated with juvenile nephronophthisis is present in a subset of individuals with joubert syndrome. Am J Hum Genet. 2004;75:82–91.
Senocak EU, Oguz KK, Haliloglu G, Topcu M, Cila A. Structural abnormalities of the brain other than molar tooth sign in joubert syndrome-related disorders. Diagn Interv Radiol. 2010;16:3–6.
Koyama S, Sato H, Wada M, Kawanami T, Emi M, Kato T. Whole-exome sequencing and digital pcr identified a novel compound heterozygous mutation in the nphp1 gene in a case of joubert syndrome and related disorders. BMC Med Genet. 2017;18:37.
Mackie FL, Carss KJ, Hillman SC, Hurles ME, Kilby MD. Exome sequencing in fetuses with structural malformations. J Clin Med. 2014;3:747–62.
Pangalos C, Hagnefelt B, Lilakos K, Konialis C. First applications of a targeted exome sequencing approach in fetuses with ultrasound abnormalities reveals an important fraction of cases with associated gene defects. PeerJ. 2016;4:e1955.
Committee Opinion no. 682. Microarrays and next-generation sequencing technology: The use of advanced genetic diagnostic tools in obstetrics and gynecology. Obstet Gynecol. 2016;128:e262–e268.
ACMG Board of Directors. Points to consider in the clinical application of genomic sequencing. Genet Med. 2012;14:759–61.
Kalynchuk EJ, Althouse A, Parker LS, Saller DN Jr., Rajkovic A. Prenatal whole-exome sequencing: parental attitudes. Prenat Diagn. 2015;35:1030–6.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Reches, A., Hiersch, L., Simchoni, S. et al. Whole-exome sequencing in fetuses with central nervous system abnormalities. J Perinatol 38, 1301–1308 (2018). https://doi.org/10.1038/s41372-018-0199-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41372-018-0199-3
This article is cited by
-
Prenatal Genome-Wide Sequencing analysis (Exome or Genome) in detecting pathogenic Single Nucleotide Variants in fetal Central Nervous System Anomalies: systematic review and meta-analysis
European Journal of Human Genetics (2024)
-
Insights on the Role of α- and β-Tubulin Isotypes in Early Brain Development
Molecular Neurobiology (2023)
-
VRK1 functional insufficiency due to alterations in protein stability or kinase activity of human VRK1 pathogenic variants implicated in neuromotor syndromes
Scientific Reports (2019)
-
Subchromosomal anomalies in small for gestational-age fetuses and newborns
Archives of Gynecology and Obstetrics (2019)