Abstract
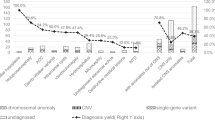
Prenatal Exome (pES) or Genome (pGS) Sequencing analysis showed a significant incremental diagnostic yield over karyotype and chromosomal microarray analysis (CMA) in fetal structural anomalies. Optimized indications and detection rates in different fetal anomalies are still under investigation. The aim of this study was to assess the incremental diagnostic yield in prenatally diagnosed Central Nervous System (CNS) anomalies. A systematic review on antenatal CNS anomalies was performed according to PRISMA guidelines, including n = 12 paper, accounting for 428 fetuses. Results were pooled in a meta‐analysis fitting a logistic random mixed-effect model. The effect of interest was the incremental diagnostic rate of pES over karyotype/CMA in detecting likely pathogenic/pathogenic Single Nucleotide Variants (SNVs). A further meta-analysis adding the available pGS studies (including diagnostic coding SNVs only) and submeta-analysis on three CNS subcategories were also performed. The pooled incremental diagnostic yield estimate of pES studies was 38% (95% C.I.: [29%;47%]) and 36% (95% C.I.: [28%;45%]) when including diagnostic SNVs of pGS studies. The point estimate of the effect resulted 22% (95% C.I.: [15%;31%]) in apparently isolated anomalies, 33% (95% C.I.: [22%;46%]) in CNS-only related anomalies (≥1) and 46% (95% C.I.: [38%;55%]) in non-isolated anomalies (either ≥ 2 anomalies in CNS, or CNS and extra-CNS). Meta-analysis showed a substantial diagnostic improvement in performing Prenatal Genome-Wide Sequencing analysis (Exome or Genome) over karyotype and CMA in CNS anomalies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data will be available upon reasonable request at the corresponding author.
References
Barbitoff YA, Polev DE, Glotov AS, Serebryakova EA, Shcherbakova IV, Kiselev AM, et al. Systematic dissection of biases in whole-exome and whole-genome sequencing reveals major determinants of coding sequence coverage. Sci Rep. 2020;10:2057. https://doi.org/10.1038/s41598-020-59026-y.
Liu P, Vossaert L. Emerging technologies for prenatal diagnosis: the application of whole genome and RNA sequencing. Prenat Diagn. 2022;42:686–96. https://doi.org/10.1002/pd.6146.
Manickam K, McClain MR, Demmer LA, Biswas S, Kearney HM, Malinowski J, et al. Exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability: an evidence-based clinical guideline of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2021;23:2029–37. https://doi.org/10.1038/s41436-021-01242-6.
Guadagnolo D, Mastromoro G, Di Palma F, Pizzuti A, Marchionni E. Prenatal exome sequencing: background, current practice and future perspectives-a systematic review. Diagnostics. 2021;11:224. https://doi.org/10.3390/diagnostics11020224.
Van den Veyver IB, Chandler N, Wilkins-Haug LE, Wapner RJ, Chitty LS, ISPD Board of Directors. International Society for Prenatal Diagnosis Updated Position Statement on the use of genome-wide sequencing for prenatal diagnosis. Prenat Diagn. 2022;42:796–803. https://doi.org/10.1002/pd.6157.
Wang Y, Greenfeld E, Watkins N, Belesiotis P, Zaidi SH, Marshall C, et al. Diagnostic yield of genome sequencing for prenatal diagnosis of fetal structural anomalies. Prenat Diagn. 2022;42:822–30. https://doi.org/10.1002/pd.6108.
Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. https://doi.org/10.1136/bmj.n71.
Munn Z, Barker TH, Moola S, Tufanaru C, Stern C, McArthur A, et al. Methodological quality of case series studies: an introduction to the JBI critical appraisal tool. JBI Evid Synth. 2020;18:2127–33. https://doi.org/10.11124/JBISRIR-D-19-00099.
Yaron Y, Ofen Glassner V, Mory A, Zunz Henig N, Kurolap A, Bar Shira A, et al. Exome sequencing as first-tier test for fetuses with severe central nervous system structural anomalies. Ultrasound Obstet Gynecol. 2022;60:59–67. https://doi.org/10.1002/uog.24885.
Lei TY, She Q, Fu F, Zhen L, Li R, Yu QX, et al. Prenatal exome sequencing in fetuses with callosal anomalies. Prenat Diagn. 2022;42:744–52. https://doi.org/10.1002/pd.6107.
de Koning MA, Hoffer MJV, Nibbeling EAR, Bijlsma EK, Toirkens MJP, Adama-Scheltema PN, et al. Prenatal exome sequencing: a useful tool for the fetal neurologist. Clin Genet. 2022;101:65–77. https://doi.org/10.1111/cge.14070.
She Q, Tang E, Peng C, Wang L, Wang D, Tan W. Prenatal genetic testing in 19 fetuses with corpus callosum abnormality. J Clin Lab Anal. 2021;35:e23971. https://doi.org/10.1002/jcla.23971.
Heide S, Spentchian M, Valence S, Buratti J, Mach C, Lejeune E, et al. Prenatal exome sequencing in 65 fetuses with abnormality of the corpus callosum: contribution to further diagnostic delineation. Genet Med. 2020;22:1887–91. https://doi.org/10.1038/s41436-020-0872-8.
Tan H, Xie Y, Chen F, Chen M, Yu L, Chen D, et al. Novel and recurrent variants identified in fetuses with central nervous system abnormalities by trios-medical exome sequencing. Clin Chim Acta. 2020;510:599–604. https://doi.org/10.1016/j.cca.2020.08.018.
Li L, Fu F, Li R, Xiao W, Yu Q, Wang D, et al. Genetic tests aid in counseling of fetuses with cerebellar vermis defects. Prenat Diagn. 2020;40:1228–38. https://doi.org/10.1002/pd.5732.
Weitensteiner V, Zhang R, Bungenberg J, Marks M, Gehlen J, Ralser DJ, et al. Exome sequencing in syndromic brain malformations identifies novel mutations in ACTB, and SLC9A6, and suggests BAZ1A as a new candidate gene. Birth Defects Res. 2018;110:587–97. https://doi.org/10.1002/bdr2.1200.
Reches A, Hiersch L, Simchoni S, Barel D, Greenberg R, Ben Sira L, et al. Whole-exome sequencing in fetuses with central nervous system abnormalities. J Perinatol. 2018;38:1301–8. https://doi.org/10.1038/s41372-018-0199-3.
Poirier K, Martinovic J, Laquerrière A, Cavallin M, Fallet-Bianco C, Desguerre I, et al. Rare ACTG1 variants in fetal microlissencephaly. Eur J Med Genet. 2015;58:416–8. https://doi.org/10.1016/j.ejmg.2015.06.006.
Liao Y, Yang Y, Wen H, Wang B, Zhang T, Li S. Abnormal Sylvian fissure at 20–30 weeks as indicator of malformations of cortical development: role of prenatal whole-genome sequencing. Ultrasound Obstet Gynecol. 2022;59:552–5. https://doi.org/10.1002/uog.24771.
Yang Y, Zhao S, Sun G, Chen F, Zhang T, Song J, et al. Genomic architecture of fetal central nervous system anomalies using whole-genome sequencing. NPJ Genom Med. 2022;7:31 https://doi.org/10.1038/s41525-022-00301-4. Published 2022 May 13.
Baujat B, Mahé C, Pignon JP, Hill C. A graphical method for exploring heterogeneity in meta-analyses: application to a meta-analysis of 65 trials. Stat Med. 2002;21:2641–52. https://doi.org/10.1002/sim.1221.
Balduzzi S, Rücker G, Schwarzer G. How to perform a meta-analysis with R: a practical tutorial. Evid Based Ment Health. 2019;22:153–60. https://doi.org/10.1136/ebmental-2019-300117.
Harrer M, Cuijpers, Furukawa T, Ebert DD. Dmetar: Companion R package for the guide “Doing Meta-Analysis in R”. R package version 0.0.9000. 2019. http://dmetar.protectlab.org/.
Lord J, McMullan DJ, Eberhardt RY, Rinck G, Hamilton SJ, Quinlan-Jones E, et al. Prenatal exome sequencing analysis in fetal structural anomalies detected by ultrasonography (PAGE): a cohort study. Lancet. 2019;393:747–57. https://doi.org/10.1016/S0140-6736(18)31940-8.
Petrovski S, Aggarwal V, Giordano JL, Stosic M, Wou K, Bier L, et al. Whole-exome sequencing in the evaluation of fetal structural anomalies: a prospective cohort study. Lancet. 2019;393:758–67. https://doi.org/10.1016/S0140-6736(18)32042-7.
Mellis R, Oprych K, Scotchman E, Hill M, Chitty LS. Diagnostic yield of exome sequencing for prenatal diagnosis of fetal structural anomalies: a systematic review and meta-analysis. Prenat Diagn. 2022;42:662–85. https://doi.org/10.1002/pd.6115.
International Society for Prenatal Diagnosis; Society for Maternal and Fetal Medicine; Perinatal Quality Foundation. Joint Position Statement from the International Society for Prenatal Diagnosis (ISPD), the Society for Maternal Fetal Medicine (SMFM), and the Perinatal Quality Foundation (PQF) on the use of genome-wide sequencing for fetal diagnosis. Prenat Diagn. 2018;38:6–9. https://doi.org/10.1002/pd.5195.
Monaghan KG, Leach NT, Pekarek D, Prasad P, Rose NC, ACMG Professional Practice and Guidelines Committee. The use of fetal exome sequencing in prenatal diagnosis: a points to consider document of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2020;22:675–80. https://doi.org/10.1038/s41436-019-0731-7.
Mustafa HJ, Barbera JP, Sambatur EV, Pagani G, Yaron Y, Baptiste CD, et al. Diagnostic yield of exome sequencing in prenatal agenesis of corpus callosum: a systematic review and meta-analysis. Ultrasound Obstet Gynecol. 2023. https://doi.org/10.1002/uog.27440.
Hart AR, Vasudevan C, Griffiths PD, Foulds N, Piercy H, de Lacy P, et al. Antenatal counselling for prospective parents whose fetus has a neurological anomaly: part 2, risks of adverse outcome in common anomalies. Dev Med Child Neurol. 2022;64:23–39. https://doi.org/10.1111/dmcn.15043.
Mone F, Abu Subieh H, Doyle S, Hamilton S, Mcmullan DJ, Allen S, et al. Evolving fetal phenotypes and clinical impact of progressive prenatal exome sequencing pathways: cohort study. Ultrasound Obstet Gynecol. 2022;59:723–30. https://doi.org/10.1002/uog.24842.
Giordano JL, Wapner RJ. The fetal sequencing consortium: the value of multidisciplinary dialog and collaboration. Prenat Diagn. 2022;42:807–10. https://doi.org/10.1002/pd.6190.
Chandler NJ, Scotchman E, Mellis R, Ramachandran V, Roberts R, Chitty LS. Lessons learnt from prenatal exome sequencing. Prenat Diagn. 2022;42:831–44. https://doi.org/10.1002/pd.6165.
Miller DT, Lee K, Abul-Husn NS, Amendola LM, Brothers K, Chung WK, et al. ACMG SF v3.2 list for reporting of secondary findings in clinical exome and genome sequencing: a policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2023;25:100866. https://doi.org/10.1016/j.gim.2023.100866.
Brew CE, Castro BA, Pan V, Hart A, Blumberg B, Wicklund C. Genetics professionals’ attitudes toward prenatal exome sequencing. J Genet Couns. 2019;28:229–39. https://doi.org/10.1002/jgc4.1112.
Vaknin N, Azoulay N, Tsur E, Tripolszki K, Urzi A, Rolfs A, et al. High rate of abnormal findings in Prenatal Exome Trio in low risk pregnancies and apparently normal fetuses. Prenat Diagn. 2022;42:725–35. https://doi.org/10.1002/pd.6077.
Wang H, Dong Z, Zhang R, Chau MHK, Yang Z, Tsang KYC, et al. Low-pass genome sequencing versus chromosomal microarray analysis: implementation in prenatal diagnosis. Genet Med. 2020;22:500–10. https://doi.org/10.1038/s41436-019-0634-7.
Stranneheim H, Lagerstedt-Robinson K, Magnusson M, Kvarnung M, Nilsson D, Lesko N, et al. Integration of whole genome sequencing into a healthcare setting: high diagnostic rates across multiple clinical entities in 3219 rare disease patients. Genome Med 2021;13:40. https://doi.org/10.1186/s13073-021-00855-5.
Acknowledgements
We acknowledge Edoardo Marchionni*, for the contribution to the statistical methodology and formal meta-analysis. *Mathematical Engineering, Statistical Learning, Department of Mathematics, Politecnico di Milano University, Milan, Italy.
Funding
The study received no funding.
Author information
Authors and Affiliations
Contributions
Study conceptualization, EM; data curation EM, DG, GM, formal analysis, EM; funding acquisition N/A; Investigation N/A; methodology, EM; DG; GM; project administration EM, AP; Resources: EM, DG, GM; writing—original draft preparation EM; DG, writing—review and editing EM, DG; GM; AP; manuscript supervision AP.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Marchionni, E., Guadagnolo, D., Mastromoro, G. et al. Prenatal Genome-Wide Sequencing analysis (Exome or Genome) in detecting pathogenic Single Nucleotide Variants in fetal Central Nervous System Anomalies: systematic review and meta-analysis. Eur J Hum Genet (2024). https://doi.org/10.1038/s41431-024-01590-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41431-024-01590-2