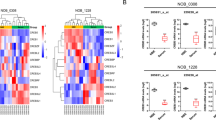
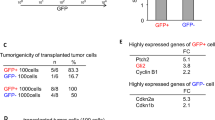
Abstract
Glioblastoma can originate from terminally differentiated astrocytes and neurons, which can dedifferentiate to a stem cell-like state upon transformation. In this study, we confirmed that transformed dedifferentiated astrocytes and neurons acquired a stem/progenitor cell state, although they still retained gene expression memory from their parental cell. Transcriptional network analysis on these cells identified upregulated genes in three main pathways: Wnt signaling, cell cycle and focal adhesion with the gene Spp1, also known as osteopontin (OPN) serving as a key common node connecting these three pathways. Inhibition of OPN blocked the formation of neurospheres, affected the proliferative capacity of transformed neurons and reduced the expression levels of neural stem cell markers. Specific inhibition of OPN in both murine and human glioma tumors prolonged mice survival. We conclude that OPN is an important player in dedifferentiation of cells during tumor formation, hence its inhibition can be a therapeutic target for glioblastoma.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 1994; 367: 645–648.
Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T et al. Identification of human brain tumour initiating cells. Nature 2004; 432: 396–401.
Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F et al. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 2013; 154: 61–74.
Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O et al. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science 2012; 338: 1080–1084.
Ischenko I, Zhi J, Moll UM, Nemajerova A, Petrenko O . Direct reprogramming by oncogenic Ras and Myc. Proc Natl Acad Sci USA 2013; 110: 3937–3942.
Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Goktuna SI, Ziegler PK et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 2013; 152: 25–38.
Bhargava V, Ko P, Willems E, Mercola M, Subramaniam S . Quantitative transcriptomics using designed primer-based amplification. Sci Rep 2013; 3: 1740.
Zhao W, Ji X, Zhang F, Li L, Ma L . Embryonic stem cell markers. Molecules 2012; 17: 6196–6236.
Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci 2008; 28: 264–278.
Polakis P . Wnt signaling in cancer. Cold Spring Harb Perspect Biol 2012; 4: a008052.
Holland JD, Klaus A, Garratt AN, Birchmeier W . Wnt signaling in stem and cancer stem cells. Curr Opin Cell Biol 2013; 25: 254–264.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Wang DD, Bordey A . The astrocyte odyssey. Prog Neurobiol 2008; 86: 342–367.
Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res 2003; 31: 374–378.
Chatr-Aryamontri A, Breitkreutz BJ, Heinicke S, Boucher L, Winter A, Stark C et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res 2013; 41: D816–D823.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13: 2498–2504.
Jan HJ, Lee CC, Shih YL, Hueng DY, Ma HI, Lai JH et al. Osteopontin regulates human glioma cell invasiveness and tumor growth in mice. Neuro Oncol 2010; 12: 58–70.
Mason CK, McFarlane S, Johnston PG, Crowe P, Erwin PJ, Domostoj MM et al. Agelastatin A: a novel inhibitor of osteopontin-mediated adhesion, invasion, and colony formation. Mol Cancer Ther 2008; 7: 548–558.
Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 2009; 5: 111–123.
Ghosh Z, Wilson KD, Wu Y, Hu S, Quertermous T, Wu JC . Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PloS One 2010; 5: e8975.
Marchetto MC, Yeo GW, Kainohana O, Marsala M, Gage FH, Muotri AR . Transcriptional signature and memory retention of human-induced pluripotent stem cells. PloS One 2009; 4: e7076.
Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol 2011; 13: 541–549.
Friedmann-Morvinski D, Verma IM . Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO Rep 2014; 15: 244–253.
Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 2008; 40: 499–507.
Polakis P . Wnt signaling and cancer. Genes Dev 2000; 14: 1837–1851.
Malumbres M, Barbacid M . Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 2009; 9: 153–166.
Denhardt DT, Noda M, O'Regan AW, Pavlin D, Berman JS . Osteopontin as a means to cope with environmental insults: regulation of inflammation, tissue remodeling, and cell survival. J Clin Invest 2001; 107: 1055–1061.
Wai PY, Kuo PC . The role of Osteopontin in tumor metastasis. J Surg Res 2004; 121: 228–241.
Katagiri YU, Sleeman J, Fujii H, Herrlich P, Hotta H, Tanaka K et al. CD44 variants but not CD44s cooperate with beta1-containing integrins to permit cells to bind to osteopontin independently of arginine-glycine-aspartic acid, thereby stimulating cell motility and chemotaxis. Cancer Res 1999; 59: 219–226.
Weber GF, Ashkar S, Glimcher MJ, Cantor H . Receptor-ligand interaction between CD44 and osteopontin (Eta-1). Science 1996; 271: 509–512.
Weber GF . The metastasis gene osteopontin: a candidate target for cancer therapy. Biochim Biophys Acta 2001; 1552: 61–85.
Rangaswami H, Bulbule A, Kundu GC . Osteopontin: role in cell signaling and cancer progression. Trends Cell Biol 2006; 16: 79–87.
Bache M, Kappler M, Wichmann H, Rot S, Hahnel A, Greither T et al. Elevated tumor and serum levels of the hypoxia-associated protein osteopontin are associated with prognosis for soft tissue sarcoma patients. BMC Cancer 2010; 10: 132.
Sreekanthreddy P, Srinivasan H, Kumar DM, Nijaguna MB, Sridevi S, Vrinda M et al. Identification of potential serum biomarkers of glioblastoma: serum osteopontin levels correlate with poor prognosis. Cancer Epidemiol Biomarkers Prev 2010; 19: 1409–1422.
Atai NA, Bansal M, Lo C, Bosman J, Tigchelaar W, Bosch KS et al. Osteopontin is up-regulated and associated with neutrophil and macrophage infiltration in glioblastoma. Immunology 2011; 132: 39–48.
Lamour V, Le Mercier M, Lefranc F, Hagedorn M, Javerzat S, Bikfalvi A et al. Selective osteopontin knockdown exerts anti-tumoral activity in a human glioblastoma model. Int J Cancer 2010; 126: 1797–1805.
Yamaguchi Y, Shao Z, Sharif S, Du XY, Myles T, Merchant M et al. Thrombin-cleaved fragments of osteopontin are overexpressed in malignant glial tumors and provide a molecular niche with survival advantage. J Biol Chem 2013; 288: 3097–3111.
Pietras A, Katz AM, Ekstrom EJ, Wee B, Halliday JJ, Pitter KL et al. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell 2014; 14: 357–369.
Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006; 9: 157–173.
McCarthy KD, de Vellis J . Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol 1980; 85: 890–902.
Meyer-Franke A, Kaplan MR, Pfrieger FW, Barres BA . Characterization of the signaling interactions that promote the survival and growth of developing retinal ganglion cells in culture. Neuron 1995; 15: 805–819.
Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D et al. A third-generation lentivirus vector with a conditional packaging system. J Virol 1998; 72: 8463–8471.
Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol 2011; 29: 79–83.
Ikawa M, Tanaka N, Kao WW, Verma IM . Generation of transgenic mice using lentiviral vectors: a novel preclinical assessment of lentiviral vectors for gene therapy. Mol Ther 2003; 8: 666–673.
Bhargava V, Head SR, Ordoukhanian P, Mercola M, Subramaniam S . Technical variations in low-input RNA-seq methodologies. Sci Rep 2014; 4: 3678.
Langmead B . Aligning short sequencing reads with Bowtie. Current Protoc Bioinformatics 2010 (Chapter 11): Unit 11 17.
Jain N, Thatte J, Braciale T, Ley K, O'Connell M, Lee JK . Local-pooled-error test for identifying differentially expressed genes with a small number of replicated microarrays. Bioinformatics 2003; 19: 1945–1951.
Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell 2002; 1: 269–277.
Acknowledgements
We thank Gabriela Estepa for her technical help and Ruben Alvarez Rodriguez for designing the inducible miRNA lentivector. IMV is an American Cancer Society Professor of Molecular Biology, and holds the Irwin and Joan Jacobs Chair in Exemplary Life Science. This work was supported in part by grants from the NIH (HL053670) (to IM Verma and D Friedmann-Morvinski), Cancer Center Core Grant (P30 CA014195-38), the H.N. and Frances C. Berger Foundation, and the Leona M. and Harry B. Helmsley Charitable Trust grant #2012-PG-MED002 (to IM Verma). S Subramaniam is supported by National Institutes of Health (NIH) National Heart, Lung and Blood Institute Grant HL087375, HL106579, HL108735, the National Science Foundation (NSF) grants DBI-0835541 and STC-0939370. V Bhargava was the recipient of a California Institute for Regenerative Medicine (CIRM) Graduate Fellowship (T1-00003 and TG2-01154, Interdisciplinary Stem Cell Training Program at UCSD). Accession Code. Gene Expression Omnibus: GSE64411 (sequencing read data).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Friedmann-Morvinski, D., Bhargava, V., Gupta, S. et al. Identification of therapeutic targets for glioblastoma by network analysis. Oncogene 35, 608–620 (2016). https://doi.org/10.1038/onc.2015.119
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.119
This article is cited by
-
P32-specific CAR T cells with dual antitumor and antiangiogenic therapeutic potential in gliomas
Nature Communications (2021)
-
Tenascin C promotes cancer cell plasticity in mesenchymal glioblastoma
Oncogene (2020)
-
The Retina of Osteopontin deficient Mice in Aging
Molecular Neurobiology (2018)