Key Points
-
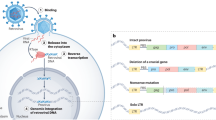
Retroviral infection of germ cells can result in virus endogenization. Up to 10% of the vertebrate genome now comprises DNA derived from such germline invaders.
-
Analysis of such endogenous retroviruses (ERVs) shows that vertebrates have been subjected to multiple waves of infection by exogenous retroviruses, with essentially the same structures as modern viruses, over a period spanning many tens of millions of years.
-
Retroviral inheritance can have both positive and negative effects on hosts. Beneficial effects include the provision of functions necessary for placenta formation and resistance to novel retrovirus infection, whereas detrimental effects include tumour induction and (presumably) genome instability.
-
Control of retrovirus expression to protect against the negative effects of retrovirus replication seems to be of considerable importance; consequently, a range of systems for blocking virus replication have been developed, including epigenetic silencing and the evolution of specific virus restriction factors.
-
Evolutionary studies indicate that an arms race between viruses and hosts has taken place, with the development of a number of viral strategies to outwit host defences. Changes to virus and host are continuing to the present day.
-
Analysis of these interactions will greatly enhance our understanding of virus replication and may suggest novel therapeutic approaches to antiretroviral drug design.
Abstract
Retroviral replication involves the formation of a DNA provirus integrated into the host genome. Through this process, retroviruses can colonize the germ line to form endogenous retroviruses (ERVs). ERV inheritance can have multiple adverse consequences for the host, some resembling those resulting from exogenous retrovirus infection but others arising by mechanisms unique to ERVs. Inherited retroviruses can also confer benefits on the host. To meet the different threats posed by endogenous and exogenous retroviruses, various host defences have arisen during evolution, acting at various stages on the retrovirus life cycle. In this Review, I describe our current understanding of the distribution and architecture of ERVs, the consequences of their acquisition for the host and the emerging details of the intimate evolutionary relationship between virus and vertebrate host.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Coffin, J. M., Hughes, S. H. & Varmus, H. E. (eds) Retroviruses (Cold Spring Harbor Press, 1997).
Kurth, R. & Bannert, N. (eds) Retroviruses: Molecular Biology, Genomics and Pathogenesis (eds Kurth, R. & Bannert, N.) (Caister Academic, 2010).
Goff, S. P. in Fields Virology Ch. 55 (eds Knipe, D. M. et al.) 1999–2069 (Lippincott Williams & Wilkins, 2007).
Hirsch, A. J. The use of RNAi-based screens to identify host proteins involved in viral replication. Future Microbiol. 5, 303–311 (2010).
Boeke, J. D. & Stoye, J. P. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. Retroviruses [online], (1997). A good introduction to the basic properties of ERVs.
Weiss, R. A. The discovery of endogenous retroviruses. Retrovirology 3, 67 (2006). A personal account of the history of ERV discovery by one of the main players in the field.
Rosenberg, N. & Jolicoeur, P. in Retroviruses (eds Coffin, J. M., Hughes, S. H. & Varmus, H. E.) 475–585 (Cold Spring Harbor Press, 1997).
Martin, M. A., Bryan, T., Rasheed, S. & Khan, A. S. Identification and cloning of endogenous retroviral sequences present in human DNA. Proc. Natl Acad. Sci. USA 78, 4892–4896 (1981).
Medstrand, P. & Blomberg, J. Characterization of novel reverse transcriptase encoding human endigenous retroviral sequences similar to type A and type B retroviruses: differential transcription in normal human tissues. J. Virol. 67, 6778–6787 (1993).
McAllister, R. M. et al. C-type virus released from cultured human rhabdomyosarcoma cells. Nature New Biol. 235, 3–6 (1972).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Mouse Genome Sequencing Consortium et al. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002).
International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432, 695–716 (2004).
Lerat, E. Identifying repeats and transposable elements in sequenced genomes: how to find your way through the dense forest of programs. Heredity 104, 520–522 (2010).
Tristem, M. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J. Virol. 74, 3715–3730 (2000). One of the first detailed studies analysing the diversity of human ERVs.
Katzourakis, A. & Tristem, M. in Retroviruses and Primate Genome Evolution (ed. Sverdlov, E. D.) 186–203 (Landes Bioscience, 2005).
Stocking, C. & Kozak, C. A. Murine endogenous retroviruses. Cell. Mol. Life Sci. 65, 3383–3398 (2008).
Huda, A., Polavarapu, N., Jordan, I. K. & McDonald, J. F. Endogenous retroviruses of the chicken genome. Biol. Direct 3, 9 (2008).
Blomberg, J., Benachenhou, F., Blikstad, V., Sperber, G. & Mayer, J. Classification and nomenclature of endogenous retroviral sequences (ERVs): problems and recommendations. Gene 448, 115–123 (2009).
Jern, P., Stoye, J. P. & Coffin, J. M. Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS Genet. 3, e183 (2007).
Benachenhou, F. et al. Evolutionary conservation of orthoretroviral long terminal repeats (LTRs) and ab initio detection of single LTRs in genomic data. PLoS ONE 4, e5179 (2009).
Copeland, N. G., Hutchison, K. W. & Jenkins, N. A. Excision of the DBA ecotropic provirus in dilute coat-color revertants of mice occurs by homologous recombination involving the viral LTRs. Cell 33, 379–387 (1983).
Katzourakis, A., Tristem, M., Pybus, O. G. & Gifford, R. J. Discovery and analysis of the first endogenous lentivirus. Proc. Natl Acad. Sci. USA 104, 6261–6265 (2007).
Gilbert, C., Maxfield, D. G., Goodman, S. M. & Feschotte, C. Parallel germline infiltration of a lentivirus in two Malagasy lemurs. PLoS Genet. 5, e10000425 (2009).
Katzourakis, A., Gifford, R. J., Tristem, M., Gilbert, M. T. P. & Pybus, O. G. Macroevolution of complex retroviruses. Science 325, 1512 (2009).
Polavarapu, N., Bowen, N. J. & McDonald, J. F. Identification, characterization and comparative genomics of chimpanzee endogenous retroviruses. Genome Biol. 7, R51 (2006).
Smit, A. F. A. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 9, 657–663 (1999).
Goodier, J. L. & Kazazian, H. H. Jr. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135, 23–35 (2008).
Holmes, E. C. The evolution of endogenous viral elements. Cell Host Microbe 10, 368–377 (2011). A review considering the evolutionary implications of endogenous viral sequences of non-retroviral origin.
Tarlington, R. E., Meers, J. & Young, P. R. Retroviral invasion of the koala genome. Nature 442, 79–81 (2006).
Jaenisch, R. Germ line integration and endogenous transmission of the exogenous Moloney leukemia virus. Proc. Natl Acad. Sci. USA 73, 1260–1264 (1976).
Salter, D. W., Smith, E. J., Hughes, S. H., Wright, S. E. & Crittenden, L. B. Transgenic chickens: insertion of retroviral genes into the chicken germ line. Virology 157, 236–240 (1987).
Lock, L. F., Keshet, E., Gilbert, D. J., Jenkins, N. A. & Copeland, N. G. Studies on the mechanism of spontaneous germline ecotropic provirus acquisition in mice. EMBO J. 7, 4169–4177 (1988).
Belshaw, R. et al. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl Acad. Sci. USA 101, 4894–4899 (2004).
Nakagawa, K. & Harrison, L. C. The potential roles of endogenous retroviruses in autoimmunity. Immunol. Rev. 152, 193–236 (1996).
Ruprecht, K., Mayer, J., Sauter, M., Roemer, K. & Mueller-Lantzsch, N. Endogenous retroviruses and cancer. Cell. Mol. Life Sci. 65, 3366–3382 (2008).
Voisset, C., Weiss, R. A. & Griffiths, D. J. Human RNA “rumor” viruses: the search for novel human retroviruses in chronic disease. Microbiol. Mol. Biol. Rev. 72, 157–196 (2008). An at times amusing account of the problems and pitfalls encountered in trying to identify possible retroviral causes for human diseases.
Li, F., Nellaker, C., Yolken, R. H. & Karlsson, H. A systematic evaluation of expression of HERV-W elements; influence of genomic context, viral structure and orientation. BMC Genomics 12, 22 (2011).
Kaufmann, S. et al. Human endogenous retrovirus protein Rec. interacts with the testicular zinc-finger protein and androgen receptor. J. Gen. Virol. 91, 1494–1502 (2010).
Lauring, A. S., Anderson, M. M. & Overbaugh, J. Specificity in receptor usage by T-cell-tropic feline leukemia viruses: implications for the in vivo tropism of immunodeficiency-inducing variants. J. Virol. 75, 8888–8898 (2001).
Stoye, J. P., Moroni, C. & Coffin, J. M. Virological events leading to spontaneous AKR thymomas. J. Virol. 65, 1273–1285 (1991).
Maksakova, I. A. et al. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2, e2 (2006).
Cohen, C. J., Lock, W. M. & Mager, D. L. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene 448, 105–114 (2009).
Lamprecht, B., Bonifer, C. & Mathas, S. Repeat-element driven activation of proto-oncogenes in human malignancies. Cell Cycle 9, 4276–4281 (2010).
Zhang, Y., Romanish, M. T. & Mager, D. L. Distributions of transposable elements reveal hazardous zones in mammalian introns. PLoS Comput. Biol. 7, e1002046 (2011).
Hughes, J. F. & Coffin, J. M. Evidence for genomic rearrangements mediated by human endogenous retroviruses during primate evolution. Nature Genet. 29, 487–489 (2001).
Romanish, M. T., Cohen, C. J. & Mager, D. L. Potential mechanisms of endogenous retroviral-mediated genomic instability in human cancer. Semin. Cancer Biol. 20, 246–253 (2010).
Sun, C. et al. Deletion of the azoospermia factor a (AZFa) region of human Y chromosome caused by recombination between HERV15 proviruses. Hum. Mol. Genet. 9, 2391–2396 (2000).
Mi, S. et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403, 785–789 (2000).
Dupressoir, A. et al. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc. Natl Acad. Sci. USA 106, 12127–12132 (2009). The first unequivocal evidence that a protein encoded by an ERV has a key role in vertebrate development.
Mangeney, M. et al. Placental syncytins: genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc. Natl Acad. Sci. USA 104, 20534–20539 (2007).
Heidmann, O., Vernochet, C., Dupressoir, A. & Heidmann, T. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: a new “syncytin” in a third order of mammals. Retrovirology 6, 107 (2009).
Meisler, M. H. & Ting, C. N. The remarkable evolutionary history of the human amylase genes. Crit. Rev. Oral Biol. Med. 4, 503–509 (1993).
Beyer, U., Moll-Rocek, J., Moll, U. M. & Dobbelstein, M. Endogenous retrovirus drives hitherto unknown proapoptotic p63 isoforms in the male germ line of humans and great apes. Proc. Natl Acad. Sci. USA 108, 3624–3629 (2011).
Walsh, C. P. & Bestor, T. H. Cytosine methylation and mammalian development. Genes Dev. 13, 26–34 (1999).
Maksakova, I. A., Mager, D. L. & Reiss, D. Keeping active endogenous retroviral-like elements in check: the epigenetic perspective. Cell. Mol. Life Sci. 65, 3329–3347 (2008).
Rowe, H. M. & Trono, D. Dynamic control of endogenous retroviruses during development. Virology 411, 273–287 (2011).
Neil, S. & Bieniasz, P. Human immunodeficiency virus, restriction factors, and interferon. J. Interferon Cytokine Res. 29, 569–580 (2009).
Bushman, F. D. et al. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 5, e1000437 (2009).
Kozak, C. A. The mouse “xenotropic” gammaretroviruses and their XPR1 receptor. Retrovirology 7, 101 (2010). A comprehensive review of the evolutionary changes occurring in the mouse receptor for the XMV.
Groudine, M., Eisenman, R. & Weintraub, H. Chromatin structure of endogenous retroviral genomes and activation by an inhibitor of DNA methylation. Nature 292, 311–317 (1981).
Jähner, D. et al. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature 298, 623–628 (1982).
Wigler, M., Levy, D. & Perucho, M. The somatic replication of DNA methylation. Cell 24, 33–40 (1981).
Yoder, J. A., Walsh, C. P. & Bestor, T. H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13, 335–340 (1997).
Wolf, D. & Goff, S. P. Embyonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458, 1201–1204 (2009).
Rowe, H. M. et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463, 237–240 (2010).
Nisole, S., Stoye, J. P. & Saïb, A. Trim family proteins: retroviral restriction and antiviral defence. Nature Rev. Microbiol. 3, 799–808 (2005).
Reik, W. Stability and flexibility of epigentic gene regulation in mammalian development. Nature 411, 425–432 (2007).
Feng, S., Jacobsen, S. E. & Reik, W. Epigenetic reprogramming in plant and animal development. Science 330, 622–627 (2010).
Leung, D. C. & Lorincz, M. C. Silencing of endogenous retroviruses: when and why do histone marks predominate? Trends Biochem. Sci. 37, 127–133 (2012).
Reiss, D. & Mager, D. L. Stochastic epigenetic silencing of retrotransposons: does stability come with age? Gene 390, 130–135 (2007).
Morgan, H. D., Sutherland, H. G., Martin, D. I. & Whitelaw, E. Epigenetic inheritance at the agouti locus in the mouse. Nature Genet. 23, 314–318 (1999).
Sheehy, A. M., Gaddis, N. C., Choi, J. D. & Malim, M. H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650 (2002). The first report of the cloning of the host resistance gene being overcome by the lentivirus accessory gene vif.
Malim, M. H. APOBEC proteins and intrinsic resistance to HIV-1 infection. Phil. Trans. R. Soc. B 364, 675–687 (2009).
Chiu, Y. L. & Greene, W. C. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 26, 317–353 (2008).
Harris, R. S. et al. DNA deamination mediates innate immunity to retroviral infection. Cell 113, 803–809 (2003).
Harris, R. S., Sheehy, A. M., Craig, H. M., Malim, M. H. & Neuberger, M. S. DNA deamination: not just a trigger for antibody diversification but also a mechanism for defense against retroviruses. Nature Immunol. 4, 641–643 (2003).
Newman, E. N. et al. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15, 166–170 (2005).
Mbisa, J. L., Bu, W. & Pathak, V. K. APOBEC3F and APOBEC3G inhibit HIV-1 DNA integration by different mechanisms. J. Virol. 84, 5250–5259 (2010).
Huthoff, H. & Towers, G. J. Restriction of retroviral replication by APOBEC3G/F and TRIM5α. Trends Microbiol. 16, 612–619 (2008).
Stremlau, M. et al. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427, 848–853 (2004). The initial description of TRIM5α as a cellular factor inhibiting HIV-1 replication.
Stremlau, M. et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc. Natl Acad. Sci. USA 103, 5514–5519 (2006).
Perez-Caballero, D., Hatziioannou, T., Yang, A., Cowan, S. & Bieniasz, P. D. Human tripartite motif 5α domains responsible for retrovirus restriction activity and specificity. J. Virol. 79, 8969–8978 (2005).
Ohkura, S., Yap, M. W., Sheldon, T. & Stoye, J. P. All three variable regions of the TRIM5α B30.2 domain can contribute to the specificity of retrovirus restriction. J. Virol. 80, 8554–8565 (2006).
Ganser-Pornillos, B. K. et al. Hexagonal assembly of a restricting TRIM5α protein. Proc. Natl Acad. Sci. USA 108, 534–539 (2011).
Forshey, B. M., von Schwedler, U., Sundquist, W. I. & Aiken, S. C. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 76, 5667–5677 (2002).
Wu, X., Anderson, J. L., Campbell, E. M., Joseph, A. M. & Hope, T. J. Proteasome inhibitors uncouple rhesus TRIM5α restriction of HIV-1 reverse transcription and infection. Proc. Natl Acad. Sci. USA 103, 7465–7470 (2006).
Campbell, E. M., Perez, O., Anderson, J. L. & Hope, T. J. Visualization of a proteasome-independent intermediate during restriction of HIV-1 by rhesus TRIM5α. J. Cell Biol. 180, 549–561 (2008).
Pertel, T. et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472, 361–365 (2011).
Neil, S. J. D., Zang, T. & Bieniasz, P. D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430 (2008). A paper providing the first description of tetherin as a factor preventing retrovirus release from the cell surface.
Martin-Serrano, J. & Neil, S. J. Host factors involved in retroviral budding and release. Nature Rev. Microbiol. 9, 519–531 (2011).
Evans, D. T., Serra-Moreno, R., Singh, R. K. & Guatelli, J. C. BST-2/tetherin: a new component of the innate immune response to enveloped viruses. Trends Microbiol. 18, 388–396 (2010).
Laguette, N. et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 (2011).
Hrecka, K. et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 (2011).
Goldstone, D. C. et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 (2011).
Bieniasz, P. D. Intrinsic immunity: a front-line defense against viral attack. Nature Immun. 5, 1109–1115 (2004).
Schoggins, J. W. & Rice, C. M. Interferon-stimulated genes and their antiviral receptor function. Curr. Opin. Virol. 1, 1–7 (2011).
Stetson, D. B., Ko, J. S., Heidmann, T. & Medzhitov, R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134, 587–598 (2008).
Rice, G. I. et al. Mutations involved in Aicardi–Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nature Genet. 41, 829–832 (2009).
Beck-Engeser, G. B., Eilat, D. & Wabl, M. An autoimmune disease prevented by anti-retroviral drugs. Retrovirology 8, 91 (2011).
Gao, G., Guo, X. & Goff, S. P. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297, 1703–1706 (2002).
Dewannieux, M., Ribet, D. & Heidmann, T. Risks linked to endogenous retroviruses for vaccine production: a general overview. Biologicals 38, 366–370 (2010).
Bieniasz, P. D. & Cullen, B. R. Multiple blocks to human immunodeficiency virus type 1 replication in rodent cells. J. Virol. 74, 9868–9877 (2000).
Sherer, N. M. et al. Evolution of a species-specific determinant within human CRM1 that regulates the post-transcriptional phases of HIV-1 replication. PLoS Pathog. 7, e1002395 (2011).
Cullen, B. R. Mechanism of action of regulatory proteins encoded by complex retroviruses. Microbiol. Rev. 56, 375–394 (1992).
Malim, M. H. & Emerman, M. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe 3, 388–398 (2008). A penetrating review considering the role of viral accessory factors in overcoming host restriction factors.
Simon, J. H. et al. The regulation of primate immunodeficiency virus infectivity by Vif is cell species restricted: a role for Vif in determining virus host range and cross-species transmission. EMBO J. 17, 1259–1267 (1998).
Yu, X. et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif–Cul5–SCF complex. Science 302, 1056–1060 (2003).
Douglas, J. L. et al. The great escape: viral strategies to counter BST-2/tetherin. PLoS Pathog. 6, e1000913 (2010).
Le Tortorec, A., Willey, S. & Neil, S. J. Antiviral inhibition of enveloped virus release by tetherin/BST-2: action and counteraction. Viruses 3, 520–540 (2011).
Kaushik, R., Zhu, X., Stranska, R., Wu, Y. & Stevenson, M. A cellular restriction dictates the permissivity of nondividing monocytes/macrophages to lentivirus and gammaretrovirus infection. Cell Host Microbe 6, 68–80 (2009).
Johnson, W. E. & Sawyer, S. L. Molecular evolution of the antiretroviral TRIM5 gene. Immunogenetics 61, 163–178 (2009). A stimulating review considering the role of positive selection in modulating the evolution of the TRIM5 gene.
Ylinen, L. M. J. et al. Isolation of an active Lv1 gene from cattle indicates that tripartite motif protein-mediated innate immunity to retroviral infection is widespread among mammals. J. Virol. 80, 7332–7338 (2006).
Tareen, S. U., Sawyer, S. L., Malik, H. S. & Emerman, M. An expanded clade of rodent Trim5 genes. Virology 385, 473–483 (2009).
Sawyer, S. L., Wu, L. I., Emerman, M. & Malik, H. S. Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc. Natl Acad. Sci. USA 102, 2832–2837 (2005).
Song, B. et al. The B30.2(SPRY) domain of retroviral restriction factor TRIM5α exhibits lineage-specific length and sequence variation in primates. J. Virol. 79, 6111–6121 (2005).
Sayah, D. M., Sokolskaja, E., Berthoux, L. & Luban, J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430, 569–573 (2004).
Stoye, J. P. & Yap, M. W. Chance favors a prepared genome. Proc. Natl Acad. Sci. USA 105, 3177–3178 (2008). Describes a series of papers providing evidence that the evolution of restriction factors is continuing.
Lim, E. S., Malik, H. S. & Emerman, M. Ancient adaptive evolution of tetherin shaped the functions of Vpu and Nef in human immunodeficiency virus and primate lentiviruses. J. Virol. 84, 7124–7134 (2010).
McNatt, M. W. et al. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 5, e1000300 (2009).
Planelles, V. SAMHD1 joins the Red Queen's court. Cell Host Microbe 16, 103–105 (2012).
Goldschmidt, V. et al. Antiretroviral activity of ancestral TRIM5α. J. Virol. 82, 2089–2096 (2008).
OhAinle, M., Kerns, J. A., Li, M. M., Malik, H. S. & Emerman, M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe 4, 249–259 (2008).
Yap, M. W., Nisole, S. & Stoye, J. P. A single amino acid change in the SPRY domain of human TRIM5α leads to HIV-1 restriction. Curr. Biol. 15, 73–78 (2005).
Meyerson, N. R. & Sawyer, S. L. Two-stepping through time: mammals and viruses. Trends Micribiol. 19, 286–294 (2011).
Odaka, T., Ikeda, H. & Akatsuka, T. Restricted expression of endogenous N-tropic XC-positive leukemia virus in hybrids between G and AKR mice: an effect of the Fv-4r gene. Int. J. Cancer 25, 757–762 (1980).
Wu, T., Yan, Y. & A., K. C. Rmcf2, a xenotropic provirus in the Asian mouse species Mus castaneus, blocks infection by mouse gammaretroviruses. J. Virol. 79, 9677–9684 (2005).
Robinson, H. L. & Lamoreux, W. F. Expression of endogenous ALV antigens and susceptibility to subgroup E ALV in three strains of chickens (endogenous avian C-type virus). Virology 69, 50–62 (1976).
McDougall, A. S. et al. Defective endogenous proviruses are expressed in feline lymphoid cells: evidence for a role in natural resistance to subgroup B feline leukemia virus. J. Virol. 68, 2151–2160 (1994).
Lilly, F. Fv-2: Identification and location of a second gene governing the spleen focus response to Friend leukemia virus in mice. J. Natl Cancer Inst. 45, 163–169 (1970).
Hilditch, L. et al. Ordered assembly of murine leukemia virus capsid protein on lipid nanotubes directs specific binding by the restriction factor, Fv1. Proc. Natl Acad. Sci. USA 108, 5771–5776 (2011).
Best, S., Le Tissier, P., Towers, G. & Stoye, J. P. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382, 826–829 (1996). The first cloning of a restriction factor revealed how ERVs could become antiviral factors.
Czarneski, J., Rassa, J. C. & Ross, S. R. Mouse mammary tumor virus and the immune system. Immunol. Res. 27, 469–480 (2003).
Frankel, W. N., Rudy, C., Coffin, J. M. & Huber, B. T. Linkage of Mls genes to endogenous mammary tumour viruses of inbred mice. Nature 349, 526–528 (1991).
Gifford, R. J. Viral evolution in deep time: lentiviruses and mammals. Trends Genet. 28, 89–100 (2012).
Plantier, J. C. et al. A new human immunodeficiency virus derived from gorillas. Nature Med. 15, 871–872 (2009).
Kirmaier, A. et al. TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 8, e1000462 (2010). Provides evidence that restriction factors act to suppress cross-species transmission and can drive virus evolution.
Kim, E. Y. et al. Human APOBEC3G-mediated editing can promote HIV-1 sequence diversification and accelerate adaptation to selective pressure. J. Virol. 84, 10402–10405 (2010).
Rein, A. Genetic fingerprinting of a retroviral gag gene suggests an important role in virus replication. Proc. Natl Acad. Sci. USA 100, 11929–11930 (2003).
Goldstone, D. C. et al. Structural and functional analysis of prehistoric lentiviruses uncovers an ancient molecular interface. Cell Host Microbe 8, 248–259 (2010).
Tristem, M., Marshall, C., Karpas, A. & Hill, F. Evolution of the primate lentivirus: evidence from vpx and vpr. EMBO J. 11, 3405–3412 (1992).
Sharova, N. et al. Primate lentiviral Vpx commandeers DDB1 to counteract a macrophage restriction. PLoS Pathog. 4, e1000057 (2008).
Ribet, D. et al. An infectious progenitor for the murine IAP retrotransposon: emergence of an intracellular genetic parasite from an an ancient retrovirus. Genome Res. 18, 597–609 (2008).
Haran-Ghera, N., Peled, A., Brightman, B. K. & Fan, H. Lymphomagenesis in AKR.Fv-1b congenic mice. Cancer Res. 53, 3433–3438 (1993).
Lee, Y. N., Malim, M. H. & Bieniasz, P. D. Hypermutation of an ancient human retrovirus by APOBEC3G. J. Virol. 82, 8762–8770 (2008).
Goodchild, N. L., Wilkinson, D. A. & Mager, D. L. Recent evolutionary expansion of a subfamily of RTVL-H human endogenous retrovirus-like elements. Virology 196, 778–788 (1993).
Cordonnier, A., Casella, J.-F. & Heidmann, T. Isolation of novel human endogenous retrovirus-like elements with foamy virus-related pol sequence. J. Virol. 69, 5890–5897 (1995).
Johnson, W. E. & Coffin, J. M. Constructing primate phylogenies from ancient retrovirus sequences. Proc. Natl Acad. Sci. USA 96, 10254–10260 (1999).
Martins, H. & Villesen, P. Improved integration time estimation of endogenous retroviruses with phylogenetic data. PLoS ONE 6, e14745 (2011).
Gifford, R. & Tristem, M. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes 26, 291–315 (2003).
Andersson, M.-L., Sjottem, E., Svineng, G. & Johansen, T. Comparative analyses of the LTRs of the ERV-H family of primatespecific, retrovirus-like elements isolated from marmoset, African green monkey, and man. Virology 234, 14–30 (1997).
Shih, A., Coutavas, E. E. & Rush, M. G. Evolutionary implications of primate endogenous retroviruses. Virology 182, 495–502 (1991).
Turner, G. et al. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 11, 1531–1535 (2001).
Stoye, J. P. Endogenous retroviruses: still active after all this time? Curr. Biol. 11, R914–R916 (2001).
Goff, S. P. Host factors exploited by retroviruses. Nature Rev. Microbiol. 5, 253–263 (2007).
Hayward, W. S., Neel, B. G. & Astrin, S. M. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature 290, 475–480 (1981).
Jenkins, N. A., Copeland, N. G., Taylor, B. A. & Lee, B. K. Dilute (d) coat colour mutation of DBA/2J mice is associated with the site of integration of an ecotropic MuLV genome. Nature 293, 370–374 (1981).
Magiorkinis, G., Gifford, R. J., Katzourakis, A., De Ranter, J. & Belshaw, R. Env-less endogenous retroviruses are genomic superspreaders. Proc. Natl Acad. Sci. USA 23 Apr 2012 (doi:10.1073/pnas.1200913109).
Acknowledgements
I thank numerous colleagues in the retrovirology community and at the National Institute for Medical Research, London, UK, for many helpful discussions, and I apologize to those whose publications could not be cited here on account of space restrictions. Work in my laboratory is supported by the UK Medical Research Council (file reference U117512710).
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Related links
Glossary
- Tumorigenic
-
Capable of forming tumours. Some but not all retroviruses are capable of changing cell growth properties, resulting in cancer. The kinds of tumours seen include carcinomas, sarcomas and leukaemias.
- Provirus
-
The DNA form of a retrovirus integrated into the genomes of retrovirus-infected cells or organisms. Coding sequences are flanked by long terminal repeats.
- siRNA screens
-
(Small interfering RNA screens). Widely used 'knockout' studies of gene function that use siRNAs, which are double-stranded RNA molecules of 20–25 nucleotides in length that are capable of interfering with the expression of RNA.
- Somatic cells
-
Differentiated cells of the body that lack potential to contribute to the germ line.
- Solo LTRs
-
(Solo long terminal repeats). Lone LTRs in the genome. Homologous recombination between the two LTRs of a provirus results in excision of most of the provirus, leaving behind a solitary LTR in the genome at the site of the previous provirus.
- Retrotransposons
-
Genetic elements that can increase in copy numbers by a mechanism involving reverse transcription of an RNA intermediate followed by integration into the genome.
- Long interspersed nuclear elements
-
(LINEs). Long retrotransposons that encode reverse transcriptases but show genetic organizations and modes of amplification that are different from those of retroviruses; in particular, they lack long terminal repeats.
- Short interspersed nuclear elements
-
(SINEs). Short retrotransposons with no coding sequences. They can be reverse transcribed by LINE-encoded reverse transcriptases.
- Polyadenylation
-
mRNAs are characterized by a stretch of adenine residues at their 3′ termini. Addition of these poly(A) tails, polyadenylation, is an important step in mRNA maturation and is signalled by specific nucleotide sequences.
- Splice acceptor
-
A sequence element that is important for RNA splicing. RNA splicing is a vital step in RNA maturation in which coding exons are joined by intron removal. It is signalled by specific splice donor and splice acceptor sequences.
- Epigenetic mechanisms
-
Means by which gene expression can be modulated without altering the primary nucleotide sequences of the genes. Examples include cytosine methylation and histone deacetylation.
Rights and permissions
About this article
Cite this article
Stoye, J. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol 10, 395–406 (2012). https://doi.org/10.1038/nrmicro2783
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2783
This article is cited by
-
Genome mining shows that retroviruses are pervasively invading vertebrate genomes
Nature Communications (2023)
-
The important biological roles of Syncytin-1 of human endogenous retrovirus W (HERV-W) and Syncytin-2 of HERV-FRD in the human placenta development
Molecular Biology Reports (2023)
-
Activation of lnc-ALVE1-AS1 inhibited ALV-J replication through triggering the TLR3 pathway in chicken macrophage like cell line
Veterinary Research Communications (2023)
-
Endogenous viral elements reveal associations between a non-retroviral RNA virus and symbiotic dinoflagellate genomes
Communications Biology (2023)
-
An endogenous lentivirus in the germline of a rodent
Retrovirology (2022)