Abstract
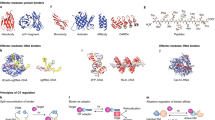
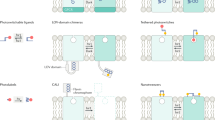
The light-based control of ion channels has been transformative for the neurosciences, but the optogenetic toolkit does not stop there. An expanding number of proteins and cellular functions have been shown to be controlled by light, and the practical considerations in deciding between reversible optogenetic systems (such as systems that use light-oxygen-voltage domains, phytochrome proteins, cryptochrome proteins and the fluorescent protein Dronpa) are well defined. The field is moving beyond proof of concept to answering real biological questions, such as how cell signalling is regulated in space and time, that were difficult or impossible to address with previous tools.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kaplan, J. H., Forbush, B. & Hoffman, J. F. Rapid photolytic release of adenosine 5′-triphosphate from a protected analogue: utilization by the Na:K pump of human red blood cell ghosts. Biochemistry 17, 1929–1935 (1978).
Callaway, E. M. & Katz, L. C. Photostimulation using caged glutamate reveals functional circuitry in living brain slices. Proc. Natl Acad. Sci. USA 90, 7661–7665 (1993).
Zemelman, B. V., Lee, G. A., Ng, M. & Miesenbock, G. Selective photostimulation of genetically chARGed neurons. Neuron 33, 15–22 (2002).
Boyden, E. S., Zhang, F., Bamberg, E., Nagel, G. & Deisseroth, K. Millisecond-timescale, genetically targeted optical control of neural activity. Nature Neurosci. 8, 1263–1268 (2005).
Li, X. et al. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc. Natl Acad. Sci. USA 102, 17816–17821 (2005).
Zhang, F. et al. Multimodal fast optical interrogation of neural circuitry. Nature 446, 633–639 (2007).
Arrenberg, A. B., Stainier, D. Y., Baier, H. & Huisken, J. Optogenetic control of cardiac function. Science 330, 971–974 (2010).
Tsai, H. C. et al. Phasic firing in dopaminergic neurons is sufficient for behavioral conditioning. Science 324, 1080–1084 (2009).
Liu, H. et al. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322, 1535–1539 (2008).
Kennedy, M. J. et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nature Methods 7, 973–975 (2010).
Bugaj, L. J., Choksi, A. T., Mesuda, C. K., Kane, R. S. & Schaffer, D. V. Optogenetic protein clustering and signaling activation in mammalian cells. Nature Methods 10, 249–252 (2013).
Christie, J. M., Salomon, M., Nozue, K., Wada, M. & Briggs, W. R. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc. Natl Acad. Sci. USA 96, 8779–8783 (1999).
Harper, S. M., Neil, L. C. & Gardner, K. H. Structural basis of a phototropin light switch. Science 301, 1541–1544 (2003).
Wu, Y. I. et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461, 104–108 (2009).
Strickland, D. et al. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nature Methods 9, 379–384 (2012).
Ni, M., Tepperman, J. M. & Quail, P. H. Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400, 781–784 (1999).
Shimizu-Sato, S., Huq, E., Tepperman, J. M. & Quail, P. H. A light-switchable gene promoter system. Nature Biotech. 20, 1041–1044 (2002).
Levskaya, A., Weiner, O. D., Lim, W. A. & Voigt, C. A. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001 (2009).
Zhou, X. X., Chung, H. K., Lam, A. J. & Lin, M. Z. Optical control of protein activity by fluorescent protein domains. Science 338, 810–814 (2012).
Ando, R., Mizuno, H. & Miyawaki, A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science 306, 1370–1373 (2004).
Fenno, L., Yizhar, O. & Deisseroth, K. The development and application of optogenetics. Annu. Rev. Neurosci. 34, 389–412 (2011).
Ryu, M. H., Moskvin, O. V., Siltberg-Liberles, J. & Gomelsky, M. Natural and engineered photoactivated nucleotidyl cyclases for optogenetic applications. J. Biol. Chem. 285, 41501–41508 (2010).
Stierl, M. et al. Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J. Biol. Chem. 286, 1181–1188 (2011).
Airan, R. D., Thompson, K. R., Fenno, L. E., Bernstein, H. & Deisseroth, K. Temporally precise in vivo control of intracellular signalling. Nature 458, 1025–1029 (2009).
Karunarathne, W. K., Giri, L., Patel, A. K., Venkatesh, K. V. & Gautam, N. Optical control demonstrates switch-like PIP3 dynamics underlying the initiation of immune cell migration. Proc. Natl Acad. Sci. USA 110, E1575–E1583 (2013).
Chen, D., Gibson, E. S. & Kennedy, M. J. A light-triggered protein secretion system. J. Cell Biol. 201, 631–640 (2013).
Crefcoeur, R. P., Yin, R., Ulm, R. & Halazonetis, T. D. Ultraviolet-B-mediated induction of protein–protein interactions in mammalian cells. Nature Commun. 4, 1779 (2013).
Yazawa, M., Sadaghiani, A. M., Hsueh, B. & Dolmetsch, R. E. Induction of protein-protein interactions in live cells using light. Nature Biotech. 27, 941–945 (2009).
Bonger, K. M., Rakhit, R., Payumo, A. Y., Chen, J. K. & Wandless, T. J. General method for regulating protein stability with light. ACS Chem. Biol. 9, 111–115 (2014).
Gambetta, G. A. & Lagarias, J. C. Genetic engineering of phytochrome biosynthesis in bacteria. Proc. Natl Acad. Sci. USA 98, 10566–10571 (2001).
Muller, K. et al. Synthesis of phycocyanobilin in mammalian cells. Chem. Commun. (Camb.) 49, 8970–8972 (2013).
Mas, P., Devlin, P. F., Panda, S. & Kay, S. A. Functional interaction of phytochrome B and cryptochrome 2. Nature 408, 207–211 (2000).
Wang, X., Chen, X. & Yang, Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nature Methods 9, 266–269 (2012).
Motta-Mena, L. B. et al. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nature Chem. Biol. 10, 196–202 (2014).
Idevall-Hagren, O., Dickson, E. J., Hille, B., Toomre, D. K. & De Camilli, P. Optogenetic control of phosphoinositide metabolism. Proc. Natl Acad. Sci. USA 109, E2316–E2323 (2012).
Wend, S. et al. Optogenetic control of protein kinase activity in mammalian cells. ACS Synth. Biol. 3, 280–285 (2013).
Leung, D. W., Otomo, C., Chory, J. & Rosen, M. K. Genetically encoded photoswitching of actin assembly through the Cdc42–WASP–Arp2/3 complex pathway. Proc. Natl Acad. Sci. USA 105, 12797–12802 (2008).
Yang, X., Jost, A. P., Weiner, O. D. & Tang, C. A light-inducible organelle targeting system for dynamically activating and inactivating signaling in budding yeast. Mol. Biol. Cell 24, 2419–2430 (2013).
Toettcher, J. E., Gong, D., Lim, W. A. & Weiner, O. D. Light-based feedback for controlling intracellular signaling dynamics. Nature Methods 8, 837–839 (2011).
Toettcher, J. E., Weiner, O. D. & Lim, W. A. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155, 1422–1434 (2013).
Konermann, S. et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature 500, 472–476 (2013).
Lungu, O. I. et al. Designing photoswitchable peptides using the AsLOV2 domain. Chem. Biol. 19, 507–517 (2012).
Tyszkiewicz, A. B. & Muir, T. W. Activation of protein splicing with light in yeast. Nature Methods 5, 303–305 (2008).
Cao, J. et al. Light-inducible activation of target mRNA translation in mammalian cells. Chem. Commun. 49, 8338–8340 (2013).
Lee, S. et al. Reversible protein inactivation by optogenetic trapping in cells. Nature Methods 11, 633–636 (2014).
Rao, M. V., Chu, P. H., Hahn, K. M. & Zaidel-Bar, R. An optogenetic tool for the activation of endogenous diaphanous-related formins induces thickening of stress fibers without an increase in contractility. Cytoskeleton 70, 394–407 (2013).
Renicke, C., Schuster, D., Usherenko, S., Essen, L. O. & Taxis, C. A LOV2 domain-based optogenetic tool to control protein degradation and cellular function. Chem. Biol. 20, 619–626 (2013).
Wang, X., He, L., Wu, Y. I., Hahn, K. M. & Montell, D. J. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nature Cell Biol. 12, 591–597 (2010).
Yoo, S. K. et al. Differential regulation of protrusion and polarity by PI3K during neutrophil motility in live zebrafish. Dev. Cell 18, 226–236 (2010).
Toettcher, J. E., Gong, D., Lim, W. A. & Weiner, O. D. Light control of plasma membrane recruitment using the Phy–PIF system. Methods Enzymol. 497, 409–423 (2011).
Spencer, S. L., Gaudet, S., Albeck, J. G., Burke, J. M. & Sorger, P. K. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature 459, 428–432 (2009).
Haber, J. E. In vivo biochemistry: physical monitoring of recombination induced by site-specific endonucleases. Bioessays 17, 609–620 (1995).
Ferrell, J. E. Jr & Machleder, E. M. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science 280, 895–898 (1998).
Shimizu, T. S., Tu, Y. & Berg, H. C. A modular gradient-sensing network for chemotaxis in Escherichia coli revealed by responses to time-varying stimuli. Mol. Syst. Biol. 6, 382 (2010).
Goentoro, L., Shoval, O., Kirschner, M. W. & Alon, U. The incoherent feedforward loop can provide fold-change detection in gene regulation. Mol. Cell 36, 894–899 (2009).
Wartlick, O. et al. Dynamics of Dpp signaling and proliferation control. Science 331, 1154–1159 (2011).
Devreotes, P. N. & Zigmond, S. H. Chemotaxis in eukaryotic cells: a focus on leukocytes and Dictyostelium. Annu. Rev. Cell Biol. 4, 649–686 (1988).
Hartline, H. K., Wagner, H. G. & Ratliff, F. Inhibition in the eye of Limulus. J. Gen. Physiol. 39, 651–673 (1956).
Purvis, J. E. & Lahav, G. Encoding and decoding cellular information through signaling dynamics. Cell 152, 945–956 (2013).
Santos, S. D., Verveer, P. J. & Bastiaens, P. I. Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nature Cell Biol. 9, 324–330 (2007).
Purvis, J. E. et al. p53 dynamics control cell fate. Science 336, 1440–1444 (2012).
Zhang, K. et al. Light-mediated kinetic control reveals the temporal effect of the Raf/MEK/ERK pathway in PC12 cell neurite outgrowth. PLoS ONE 9, e92917 (2014).
Takala, H. et al. Signal amplification and transduction in phytochrome photosensors. Nature 509, 245–248 (2014).
Kim, J. I. et al. Phytochrome phosphorylation modulates light signaling by influencing the protein–protein interaction. Plant Cell 16, 2629–2640 (2004).
Medzihradszky, M. et al. Phosphorylation of phytochrome B inhibits light-induced signaling via accelerated dark reversion in Arabidopsis. Plant Cell 25, 535–544 (2013).
Hughes, R. M., Vrana, J. D., Song, J. & Tucker, C. L. Light-dependent, dark-promoted interaction between Arabidopsis cryptochrome 1 and phytochrome B proteins. J. Biol. Chem. 287, 22165–22172 (2012).
Strickland, D. et al. Rationally improving LOV domain-based photoswitches. Nature Methods 7, 623–626 (2010).
Loewer, A., Karanam, K., Mock, C. & Lahav, G. The p53 response in single cells is linearly correlated to the number of DNA breaks without a distinct threshold. BMC Biol. 11, 114 (2013).
Burgie, E. S., Bussell, A. N., Walker J.M., Dubiel K. & Vierstra R.D. Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc. Natl Acad. Sci. USA (in the press).
Acknowledgements
The authors thank members of the Weiner laboratory for helpful discussions. This work was supported by a Genentech Fellowship (D.T.) and US National Institutes of Health (NIH) grants GM084040, GM096164 and GM109899 (O.D.W.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Tischer, D., Weiner, O. Illuminating cell signalling with optogenetic tools. Nat Rev Mol Cell Biol 15, 551–558 (2014). https://doi.org/10.1038/nrm3837
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3837
This article is cited by
-
Spatiotemporal control of RNA metabolism and CRISPR–Cas functions using engineered photoswitchable RNA-binding proteins
Nature Protocols (2024)
-
Light-driven biological actuators to probe the rheology of 3D microtissues
Nature Communications (2023)
-
Spatiotemporal dynamics of membrane surface charge regulates cell polarity and migration
Nature Cell Biology (2022)
-
Optogenetic control of RNA function and metabolism using engineered light-switchable RNA-binding proteins
Nature Biotechnology (2022)
-
Optogenetic manipulation of cell migration with high spatiotemporal resolution using lattice lightsheet microscopy
Communications Biology (2022)