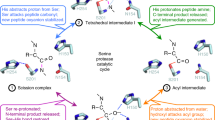
Key Points
-
Rhomboids are a newly discovered family of serine proteases that are conserved throughout evolution.
-
Unlike all other serine proteases, rhomboids do not cleave soluble substrates but instead cut protein transmembrane domains within the lipid bilayer. This is analogous to the intramembrane protein cleavage that is catalysed by presenilins, signal-peptide peptidase and site-2 protease, which are unrelated intramembrane proteases of other mechanistic classes.
-
Rhomboids were first characterized in Drosophila melanogaster, where Rhomboid-1 is the primary activator of intercellular signalling by the epidermal growth factor receptor. All rhomboids that have been studied so far release proteins in the extracellular or luminal direction; this distinguishes them from the other intramembrane proteases, almost all of which release cytoplasmic protein domains.
-
Although the biological roles of rhomboids in most organisms are not yet known, there is a subclass of mitochondrial rhomboids that, at least in yeast, controls mitochondrial membrane remodelling by cleaving a dynamin-like protein known as Mgm1. One bacterial rhomboid has been investigated and it regulates the emission of quorum-sensing signal.
-
As with the other intramembrane proteases, little is known about the catalytic mechanism of rhomboids. The highly conserved and catalytically essential residues are predicted to form a catalytic triad comparable to that in the classic soluble serine proteases, but how this operates in the context of catalysis in a lipid bilayer is uncertain. All intramembrane proteases have multiple transmembrane domains and it seems likely that these form a hydrophilic micro-environment around the catalytic centre.
-
Intramembrane proteolysis is rapidly emerging as a widespread and versatile signalling mechanism that is used to control a great variety of biological events.
Abstract
The rhomboids are a recently discovered family of serine proteases with the unusual property of cleaving proteins within their transmembrane domains. They are the most widely conserved polytopic membrane proteins discovered so far. Although not much is known about the spectrum of their biological roles, it is already clear that rhomboids control events as diverse as growth factor signalling and mitochondrial membrane dynamics. As with other intramembrane proteases, the molecular details of how proteolysis can occur in a lipid bilayer remain mysterious.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Matthews, B. W., Sigler, P. B., Henderson, R. & Blow, D. M. Three-dimensional structure of tosyl-α-chymotrypsin. Nature 214, 652–656 (1967).
Fersht, A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding (W. H. Freeman and Company, New York, 1999).
Rawson, R. B. et al. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol. Cell 1, 47–57 (1997).
Wolfe, M. S. et al. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and γ-secretase activity. Nature 398, 513–517 (1999).
Li, Y. M. et al. Photoactivated γ-secretase inhibitors directed to the active site covalently label presenilin 1. Nature 405, 689–694 (2000).
Esler, W. P. et al. Transition-state analogue inhibitors of γ-secretase bind directly to presenilin-1. Nature Cell Biol. 2, 428–434 (2000).
Weihofen, A., Binns, K., Lemberg, M. K., Ashman, K. & Martoglio, B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science 296, 2215–2218 (2002).
Urban, S., Lee, J. R. & Freeman, M. Drosophila Rhomboid-1 defines a family of putative intramembrane serine proteases. Cell 107, 173–182 (2001). This paper reports the discovery that rhomboids are a new, conserved family of intramembrane serine proteases.
Wasserman, J. D., Urban, S. & Freeman, M. A family of rhomboid-like genes: Drosophila rhomboid-1 and roughoid/rhomboid-3 cooperate to activate EGF receptor signalling. Genes Dev. 14, 1651–1663 (2000).
Guichard, A., Roark, M., Ronshaugen, M. & Bier, E. brother of rhomboid, a rhomboid-related gene expressed during early Drosophila oogenesis, promotes EGF-R/MAPK signaling. Dev. Biol. 226, 255–266 (2000).
Koonin, E. V. et al. The rhomboids: a nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers. Genome Biol. 4, R19 (2003). This paper describes a detailed phylogenetic analysis of rhomboids and points out that they are the most widely conserved polytopic membrane proteins.
Jürgens, G., Wieschaus, E., Nüsslein-Volhard, C. & Kluding, H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. II. Zygotic loci on the third chromosome. Wilhelm Roux's Arch. Dev. Biol. 193, 283–295 (1984).
Mayer, U. & Nüsslein-Volhard, C. A group of genes required for pattern formation in the ventral ectoderm of the Drosophila embryo. Genes Dev. 2, 1496–1511 (1988).
Schweitzer, R. & Shilo, B. -Z. A thousand and one roles for the Drosophila EGF receptor. Trends Genet. 13, 191–196 (1997).
Diaz-Benjumea, F. J. & Garcia-Bellido, A. Genetic analysis of the wing vein pattern in Drosophila. Wilhelm Roux's Arch. Dev. Biol. 198, 336–354 (1990).
Freeman, M., Kimmel, B. E. & Rubin, G. M. Identifying targets of the rough homeobox gene of Drosophila: evidence that rhomboid functions in eye development. Development 116, 335–346 (1992).
Bier, E., Jan, L. Y. & Jan, Y. N. rhomboid, a gene required for dorsoventral axis establishment and peripheral nervous system development in Drosophila melanogaster. Genes Dev. 4, 190–203 (1990).
Sturtevant, M. A., Roark, M. & Bier, E. The Drosophila rhomboid gene mediates the localized formation of wing veins and interacts genetically with components of the EGF-R signaling pathway. Genes Dev. 7, 961–973 (1993).
Ruohola-Baker, H. et al. Spatially localized rhomboid is required for establishment of the dorsal–ventral axis in Drosophila oogenesis. Cell 73, 953–965 (1993).
Freeman, M. The spitz gene is required for photoreceptor determination in the Drosophila eye where it interacts with the EGF receptor. Mech. Dev. 48, 25–33 (1994).
Shilo, B. Z. Signaling by the Drosophila epidermal growth factor receptor pathway during development. Exp. Cell Res. 284, 140–149 (2003).
Golembo, M., Raz, E. & Shilo, B. Z. The Drosophila embryonic midline is the site of Spitz processing, and induces activation of the EGF receptor in the ventral ectoderm. Development 122, 3363–3370 (1996).
Guichard, A. et al. rhomboid and Star interact synergistically to promote EGFR/MAPK signaling during Drosophila wing vein development. Development 126, 2663–2676 (1999). An elegant genetic demonstration that D. melanogaster Rhomboid-1 and Star function co-dependently to activate EGFR signalling.
Wasserman, J. D. & Freeman, M. An autoregulatory cascade of EGF receptor signalling patterns the Drosophila egg. Cell 95, 355–364 (1998).
zür Lage, P., Jan, Y. N. & Jarman, A. P. Requirement for EGF receptor signalling in neural recruitment during formation of Drosophila chordotonal sense organ clusters. Curr. Biol. 7, 166–175 (1997).
Brentrup, D., Lerch, H., Jackle, H. & Noll, M. Regulation of Drosophila wing vein patterning: net encodes a bHLH protein repressing rhomboid and is repressed by rhomboid-dependent Egfr signalling. Development 127, 4729–4741 (2000).
Gabay, L., Seger, R. & Shilo, B. -Z. In situ activation pattern of Drosophila EGF receptor pathway during development. Science 277, 1103–1106 (1997).
Noll, R., Sturtevant, M. A., Gollapudi, R. R. & Bier, E. New functions of the Drosophila rhomboid gene during embryonic and adult development are revealed by a novel genetic method, enhancer piracy. Development 120, 2329–2338 (1994).
Sturtevant, M. A. & Bier, E. Analysis of the genetic hierarchy guiding wing vein development in Drosophila. Development 121, 785–801 (1995).
Kolodkin, A. L., Pickup, A. T., Lin, D. M., Goodman, C. S. & Banerjee, U. Characterization of Star and its interactions with sevenless and EGF receptor during photoreceptor cell development in Drosophila. Development 120, 1731–1745 (1994).
Bang, A. G. & Kintner, C. Rhomboid and Star facilitate presentation and processing of the Drosophila TGF-α homolog Spitz. Genes Dev. 14, 177–186 (2000). The first experimental demonstration that Star and Rhomboid-1 somehow trigger the proteolytic release of Spitz.
Heberlein, U., Hariharan, I. K. & Rubin, G. M. Star is required for neuronal differentiation in the Drosophila retina and displays dosage-sensitive interactions with Ras1. Dev. Biol. 160, 51–63 (1993).
Pickup, A. T. & Banerjee, U. The role of Star in the production of an activated ligand for the EGF receptor signaling pathway. Dev. Biol. 205, 254–259 (1999).
Hsiung, F., Griffis, E. R., Pickup, A., Powers, M. A. & Moses, K. Function of the Drosophila TGF-α homolog Spitz is controlled by Star and interacts directly with Star. Mech. Dev. 107, 13–23 (2001).
Lanoue, B. R. & Jacobs, J. R. Rhomboid function in the midline of the Drosophila CNS. Dev. Genet. 25, 321–330 (1999).
Sapir, A., Schweitzer, R. & Shilo, B. -Z. Sequential activation of the EGF receptor pathway during Drosophila oogenesis establishes the dorsoventral axis. Development 125, 191–200 (1998).
Schweitzer, R., Shaharabany, M., Seger, R. & Shilo, B. -Z. Secreted Spitz triggers the DER signalling pathway and is a limiting component in embryonic ventral ectoderm determination. Genes Dev. 9, 1518–1529 (1995). Reported that the soluble form of Spitz was active but that the membrane-tethered form had no detectable activity; this led to the idea that proteolysis of Spitz is a key step in EGFR activation.
Lee, J. R., Urban, S., Garvey, C. F. & Freeman, M. Regulated intracellular ligand transport and proteolysis controls EGF signal activation in Drosophila. Cell 107, 161–171 (2001). This paper was the first to describe the mechanism by which Star and Rhomboid-1 control the trafficking and cleavage of the TGFα-like growth factor Spitz.
Ghiglione, C. et al. Mechanism of activation of the Drosophila EGF receptor by the TGFα ligand Gurken during oogenesis. Development 129, 175–186 (2002).
Tsruya, R. et al. Intracellular trafficking by Star regulates cleavage of the Drosophila EGF receptor ligand Spitz. Genes Dev. 16, 222–234 (2002).
Pascall, J. C., Luck, J. E. & Brown, K. D. Expression in mammalian cell cultures reveals interdependent, but distinct, functions for Star and Rhomboid proteins in the processing of the Drosophila transforming-growth-factor-α homologue Spitz. Biochem. J. 363, 347–352 (2002).
Pellegrini, L. et al. PAMP and PARL, two novel putative metalloproteases interacting with the COOH-terminus of Presenilin-1 and-2. J. Alzheimers Dis. 3, 181–190 (2001).
Paetzel, M. & Dalbey, R. E. Catalytic hydroxyl/amine dyads within serine proteases. Trends Biochem. Sci. 22, 28–31 (1997).
Vernet, T. et al. Structural and functional roles of asparagine 175 in the cysteine protease papain. J. Biol. Chem. 270, 16645–16652 (1995).
Ye, J., Dave, U. P., Grishin, N. V., Goldstein, J. L. & Brown, M. S. Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by site-2 protease. Proc. Natl Acad. Sci. USA 97, 5123–5128 (2000).
Mumm, J. S. et al. A ligand-induced extracellular cleavage regulates γ-secretase-like proteolytic activation of Notch1. Mol. Cell 5, 197–206 (2000).
Struhl, G. & Adachi, A. Requirements for presenilin-dependent cleavage of notch and other transmembrane proteins. Mol. Cell 6, 625–636 (2000).
Lemberg, M. K. & Martoglio, B. Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol. Cell 10, 735–744 (2002).
Sakai, J. et al. Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol. Cell 2, 505–514 (1998).
Urban, S. & Freeman, M. Substrate specificity of rhomboid intramembrane proteases is governed by helix-breaking residues in the substrate transmembrane domain. Mol. Cell 11, 1425–1434 (2003). This paper describes the requirements within the TMD of a rhomboid substrate that make it cleavable by rhomboids. It implies that destabilization of the TMD α-helix is necessary; it also predicts that a family of Toxoplasma gondii adhesion proteins are rhomboid substrates.
Soldati, D., Dubremetz, J. F. & Lebrun, M. Microneme proteins: structural and functional requirements to promote adhesion and invasion by the apicomplexan parasite Toxoplasma gondii. Int. J. Parasitol. 31, 1293–1302 (2001).
Opitz, C. et al. Intramembrane cleavage of microneme proteins at the surface of the apicomplexan parasite Toxoplasma gondii. EMBO J. 21, 1577–1585 (2002).
Urban, S., Lee, J. R. & Freeman, M. A family of Rhomboid intramembrane proteases activates all Drosophila membrane-tethered EGF ligands. EMBO J. 21, 4277–4286 (2002).
Lohi, O., Urban, S. & Freeman, M. Diverse substrate recognition mechanisms for rhomboids: thrombomodulin is cleaved by mammalian rhomboids. Curr. Biol. 14, 263–241 (2004).
Gallio, M., Sturgill, G., Rather, P. & Kylsten, P. A conserved mechanism for extracellular signaling in eukaryotes and prokaryotes. Proc. Natl Acad. Sci. USA 99, 12208–12213 (2002).
Urban, S., Schlieper, D. & Freeman, M. Conservation of intramembrane proteolytic activity and substrate specificity in prokaryotic and eukaryotic rhomboids. Curr. Biol. 12, 1507–1512 (2002).
Miller, M. B. & Bassler, B. L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199 (2001).
Rather, P. N. & Orosz, E. Characterization of aarA, a pleiotrophic negative regulator of the 2′-N-acetyltransferase in Providencia stuartii. J. Bacteriol. 176, 5140–5144 (1994).
Rather, P. N., Ding, X., Baca-DeLancey, R. R. & Siddiqui, S. Providencia stuartii genes activated by cell-to-cell signaling and identification of a gene required for production or activity of an extracellular factor. J. Bacteriol. 181, 7185–7191 (1999). References 55, 56 and 59 describe evidence that rhomboid from the bacterium Providencia stuartii controls the emission of a quorum-sensing signal. These papers provide the first evidence for shared mechanisms of intercellular signalling between bacteria and animals.
Gallio, M. & Kylsten, P. Providencia may help find a function for a novel, widespread protein family. Curr. Biol. 10, R693–R694 (2000).
Dimmer, K. S. et al. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 847–853 (2002).
McQuibban, G. A., Saurya, S. & Freeman, M. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature 423, 537–541 (2003).
Herlan, M., Vogel, F., Bornhovd, C., Neupert, W. & Reichert, A. S. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J. Biol. Chem. 278, 27781–27788 (2003). References 62 and 63 show that a mitochondrial rhomboid in yeast controls mitochondrial membrane fusion by cleaving the dynamin-like GTPase Mgm1. Reference 62 also shows that mitochondrial rhomboids are conserved in humans and other eukaryotes.
Esser, K., Tursun, B., Ingenhoven, M., Michaelis, G. & Pratje, E. A novel two-step mechanism for removal of a mitochondrial signal sequence involves the mAAA complex and the putative rhomboid protease Pcp1. J. Mol. Biol. 323, 835–843 (2002). The first report of proteolysis by a mitochondrial rhomboid in yeast. In this case the substrate was cytochrome- c peroxidase.
Shepard, K. A. & Yaffe, M. P. The yeast dynamin-like protein, Mgm1p, functions on the mitochondrial outer membrane to mediate mitochondrial inheritance. J. Cell Biol. 144, 711–720 (1999).
Wong, E. D. et al. The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J. Cell Biol. 151, 341–352 (2000).
Wong, E. D. et al. The intramitochondrial dynamin-related GTPase, Mgm1p, is a component of a protein complex that mediates mitochondrial fusion. J. Cell Biol. 160, 303–311 (2003).
Sesaki, H., Southard, S. M., Hobbs, A. E. & Jensen, R. E. Cells lacking Pcp1p/Ugo2p, a rhomboid-like protease required for Mgm1p processing, lose mtDNA and mitochondrial structure in a Dnm1p-dependent manner, but remain competent for mitochondrial fusion. Biochem. Biophys. Res. Commun. 308, 276–283 (2003).
Alexander, C. et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nature Genet. 26, 211–215 (2000).
Schulz, C., Wood, C. G., Jones, D. L., Tazuke, S. I. & Fuller, M. T. Signaling from germ cells mediated by the rhomboid homolog stet organizes encapsulation by somatic support cells. Development 129, 4523–4534 (2002).
Jaszai, J. & Brand, M. Cloning and expression of Ventrhoid, a novel vertebrate homologue of the Drosophila EGF pathway gene rhomboid. Mech. Dev. 113, 73–77 (2002).
Pascall, J. C. & Brown, K. D. Characterization of a mammalian cDNA encoding a protein with high sequence similarity to the Drosophila regulatory protein Rhomboid. FEBS Lett. 429, 337–340 (1998).
Sunnarborg, S. W. et al. Tumor necrosis factor-α converting enzyme (TACE) regulates epidermal growth factor receptor ligand availability. J. Biol. Chem. 277, 12838–12845 (2002).
Borrell-Pages, M., Rojo, F., Albanell, J., Baselga, J. & Arribas, J. TACE is required for the activation of the EGFR by TGF-α in tumors. EMBO J. 22, 1114–1124 (2003).
Peschon, J. J. et al. An essential role for ectodomain shedding in mammalian development. Science 282, 1281–1284 (1998).
Weiler, H. & Isermann, B. H. Thrombomodulin. J. Thromb. Haemost. 1, 1515–1524 (2003).
Wright, C. S., Alden, R. A. & Kraut, J. Structure of subtilisin BPN′ at 2.5 angstrom resolution. Nature 221, 235–242 (1969).
Urban, S. & Freeman, M. Intramembrane proteolysis controls diverse signaling pathways throughout evolution. Curr. Opin. Genet. Dev. 12, 512–518 (2002).
Brown, M. S., Ye, J., Rawson, R. B. & Goldstein, J. L. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100, 391–398 (2000). An influential and important review that first described the phenomenon of intramembrane proteolysis; it coined the much-used term RIP for regulated intramembrane proteolysis.
Weihofen, A. & Martoglio, B. Intramembrane-cleaving proteases: controlled liberation of proteins and bioactive peptides. Trends Cell Biol. 13, 71–78 (2003).
Huppert, S. & Kopan, R. Regulated intramembrane proteolysis takes another twist. Dev. Cell 1, 590–592 (2001).
Hua, X., Sakai, J., Ho, Y. K., Goldstein, J. L. & Brown, M. S. Hairpin orientation of sterol regulatory element-binding protein-2 in cell membranes as determined by protease protection. J. Biol. Chem. 270, 29422–29427 (1995).
Duncan, E. A., Dave, U. P., Sakai, J., Goldstein, J. L. & Brown, M. S. Second-site cleavage in sterol regulatory element-binding protein occurs at transmembrane junction as determined by cysteine panning. J. Biol. Chem. 273, 17801–17809 (1998).
Francis, R. et al. aph-1 and pen-2 are required for Notch pathway signaling, γ-secretase cleavage of βAPP, and presenilin protein accumulation. Dev. Cell 3, 85–97 (2002).
Yu, G. et al. Nicastrin modulates presenilin-mediated notch/glp-1 signal transduction and βAPP processing. Nature 407, 48–54 (2000).
De Strooper, B. Aph-1, Pen-2, and Nicastrin with Presenilin generate an active γ-Secretase complex. Neuron 38, 9–12 (2003).
Weihofen, A., Lemberg, M. K., Ploegh, H. L., Bogyo, M. & Martoglio, B. Release of signal peptide fragments into the cytosol requires cleavage in the transmembrane region by a protease activity that is specifically blocked by a novel cysteine protease inhibitor. J. Biol. Chem. 275, 30951–30956 (2000).
Edbauer, D. et al. Reconstitution of γ-secretase activity. Nature Cell Biol. 5, 486–488 (2003).
Southan, C. A genomic perspective on human proteases. FEBS Lett. 498, 214–218 (2001).
Wolfe, M. S. Secretase as a target for Alzheimer's disease. Curr. Top. Med. Chem. 2, 371–383 (2002).
DeBose-Boyd, R. A. et al. Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell 99, 703–712 (1999).
Nohturfft, A., DeBose-Boyd, R. A., Scheek, S., Goldstein, J. L. & Brown, M. S. Sterols regulate cycling of SREBP cleavage-activating protein (SCAP) between endoplasmic reticulum and Golgi. Proc. Natl Acad. Sci. USA 96, 11235–11240 (1999).
Yabe, D., Brown, M. S. & Goldstein, J. L. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc. Natl Acad. Sci. USA 99, 12753–12758 (2002).
Yang, T. et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 110, 489–500 (2002).
Ye, J. et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6, 1355–1364 (2000).
Rudner, D. Z., Fawcett, P. & Losick, R. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc. Natl Acad. Sci. USA 96, 14765–14770 (1999).
An, F. Y., Sulavik, M. C. & Clewell, D. B. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J. Bacteriol. 181, 5915–5921 (1999).
An, F. Y. & Clewell, D. B. Identification of the cAD1 sex pheromone precursor in Enterococcus faecalis. J. Bacteriol. 184, 1880–1887 (2002).
Alzheimer's Disease Collaborative Group. The structure of the presenilin 1 (S182) gene and identification of six novel mutations in early onset AD families. Nature Genet. 11, 219–222 (1995).
Selkoe, D. J. Deciphering the genesis and fate of amyloid β-protein yields novel therapies for Alzheimer disease. J. Clin. Invest. 110, 1375–1381 (2002).
De Strooper, B. et al. A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature 398, 518–522 (1999).
Struhl, G. & Greenwald, I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature 398, 522–525 (1999).
Struhl, G. & Adachi, A. Nuclear access and action of notch in vivo. Cell 93, 649–660 (1998).
Schroeter, E. H., Kisslinger, J. A. & Kopan, R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393, 382–386 (1998).
Lecourtois, M. & Schweisguth, F. Indirect evidence for Delta-dependent intracellular processing of notch in Drosophila embryos. Curr. Biol. 8, 771–774 (1998).
Martoglio, B. & Golde, T. E. Intramembrane-cleaving aspartic proteases and disease: presenilins, signal peptide peptidase and their homologs. Hum. Mol. Genet. 12 (Suppl. 2), R201–R206 (2003).
McLauchlan, J., Lemberg, M. K., Hope, G. & Martoglio, B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21, 3980–3988 (2002).
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Related links
Related links
DATABASES
Entrez
Flybase
Saccharomyces genome database
Swiss-Prot
FURTHER INFORMATION
Glossary
- METALLOPROTEASE
-
A protease that depends on a coordinated metal ion for its catalytic mechanism.
- ASPARTYL PROTEASE
-
A protease that depends on two aspartate residues that are essential for catalysis.
- SERINE PROTEASE
-
A protease that uses an activated serine as the nucleophile at the heart of its catalytic mechanism.
- POLYTOPIC MEMBRANE PROTEIN
-
A protein with several transmembrane domains.
- OXYANION
-
A catalytic intermediate of proteolysis by serine proteases; stabilization of this intermediate contributes to the overall proteolytic mechanism.
- TYPE II PROTEINS
-
Transmembrane proteins with a cytoplasmic amino terminus.
- AMPHIPATHIC
-
An α-helix in which the sequence of amino-acid residues produces distinct hydrophilic and hydrophobic faces.
- EF HAND
-
A protein motif that potentially binds calcium.
- CYTOCHROME-C PEROXIDASE
-
A yeast mitochondrial enzyme that removes potentially dangerous free radicals.
- DYNAMIN
-
A large GTPase that controls endocytosis and probably other cellular events.
- AAA PROTEASE
-
A family of ATP-dependent proteases that mediate degradation of membrane proteins; mitochondrial AAA proteases are either matrix localized (mAAA) or intermembrane-space localized (iAAA).
- ADAM FAMILY
-
(ADAM, a disintegrin and metalloprotease). A family of extracellular metalloproteases named after their characteristic ADAM domain structure.
- SREPB
-
(SREBP, sterol-response-element-binding protein). A membrane-tethered transcription factor that controls certain genes in the sterol biosynthetic pathway.
- UNFOLDED PROTEIN RESPONSE
-
An endoplasmic reticulum (ER) stress response that adapts the secretory pathway to abnormal load and protects cells from the dangers of high levels of unfolded proteins.
Rights and permissions
About this article
Cite this article
Freeman, M. Proteolysis within the membrane: rhomboids revealed. Nat Rev Mol Cell Biol 5, 188–197 (2004). https://doi.org/10.1038/nrm1334
Issue Date:
DOI: https://doi.org/10.1038/nrm1334
This article is cited by
-
Sec61β, a subunit of the Sec61 protein translocation channel at the Endoplasmic Reticulum, is involved in the transport of Gurken to the plasma membrane.
BMC Cell Biology (2009)
-
m-AAA protease-driven membrane dislocation allows intramembrane cleavage by rhomboid in mitochondria
The EMBO Journal (2007)
-
Cross genome comparisons of serine proteases in Arabidopsis and rice
BMC Genomics (2006)
-
Protease gene families in Populus and Arabidopsis
BMC Plant Biology (2006)
-
Enzyme theory holds water
Nature (2006)