Abstract
Defects in plasma membrane repair can lead to muscle and heart diseases in humans. Tripartite motif-containing protein (TRIM)72 (mitsugumin 53; MG53) has been determined to rapidly nucleate vesicles at the site of membrane damage, but the underlying molecular mechanisms remain poorly understood. Here we present the structure of Mus musculus TRIM72, a complete model of a TRIM E3 ubiquitin ligase. We demonstrated that the interaction between TRIM72 and phosphatidylserine-enriched membranes is necessary for its oligomeric assembly and ubiquitination activity. Using cryogenic electron tomography and subtomogram averaging, we elucidated a higher-order model of TRIM72 assembly on the phospholipid bilayer. Combining structural and biochemical techniques, we developed a working molecular model of TRIM72, providing insights into the regulation of RING-type E3 ligases through the cooperation of multiple domains in higher-order assemblies. Our findings establish a fundamental basis for the study of TRIM E3 ligases and have therapeutic implications for diseases associated with membrane repair.
Similar content being viewed by others
Main
Cells require the repair of plasma membrane injuries to protect the cytoplasm from external environments. In muscle tissue, plasma membrane injuries frequently occur in response to mechanical and metabolic stresses1. Fortunately, membrane-repair machinery can rapidly reseal the damaged plasma membrane2,3. Defects in membrane repair can cause diseases, including muscular dystrophy and neurodegenerative diseases4,5,6. Since the initial discovery of membrane resealing7, several membrane-repair models have been proposed: patching, constriction, exocytosis–endocytosis-mediated and endosomal sorting complex required for transport (ESCRT)-mediated models8,9. However, despite the expanding biological and biochemical understanding gained from these models in recent years10,11,12,13,14,15, the lack of structural evidence makes it difficult to elucidate the molecular mechanisms of membrane resealing.
One key factor is that extracellular Ca2+ influx triggers membrane repair at damaged sites rich in phosphatidylserine (PS)2,16 through a Ca2+-dependent ESCRT system and Ca2+-induced exocytosis of lysosomes7,17. However, despite the importance of Ca2+ influx, there is also a unique Ca2+-independent membrane-repair process known to be mediated by tripartite motif-containing protein (TRIM)72 (ref. 18). TRIM72, also known as MG53, which is highly expressed in muscle18,19, is a key initiator of the plasma membrane-repair machinery that facilitates vesicle transport to the injury site18. Additionally, TRIM72 exerts protective effects in wound healing following ischemia–reperfusion injury20 and acute kidney and lung injury21,22. It has been shown to have therapeutic potential for various injuries in both muscle and nonmuscle tissues23. In addition to participating in the membrane-repair process, TRIM72 has functions related to the innate immune response24, insulin resistance25,26 and muscle differentiation19. However, due to the lack of structural and biochemical studies, its molecular mechanism remains largely unknown.
TRIM72 is a member of the TRIM superfamily27, a major subfamily of Really Interesting New Gene (RING)-type E3 ubiquitin (Ub) ligases28. Ubiquitination is a key protein quality-control system in which Ub is attached to substrates in a reaction catalyzed by an enzymatic cascade involving E1, E2 and E3 enzymes, leading to degradation of the substrate by the 26S proteasome. In addition, Ub ligases are known to trigger signal transduction, membrane trafficking and DNA damage repair processes29. They are classified into three types based on their mechanism of Ub transfer: RING, Homologous to the E6AP C-Terminus (HECT) and RING-between-RING (RBR) ligases30. RING-type ligases are the most abundant, and TRIM RING ligases contain over 70 members among the 600 Ub ligases in humans31,32. TRIMs are characterized by multimodular domains, including the conserved RING–B-box–coiled-coil (RBCC) and variable C-terminal domains. Although their domain architecture is similar, their functions and related diseases, such as antiviral defense, cancers and inherited disorders, are extremely broad32,33,34. However, despite biological evidence, in-depth structural and biochemical research on TRIMs is quite limited due to their flexibility and complexity35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52.
Here, we have elucidated the structures of TRIM72 and higher-order assemblies of TRIM72 complexed with a phospholipid membrane using integrative structural techniques. These structures enable us to understand how TRIM72 recognizes negatively charged phospholipid domains independent of Ca2+ during membrane repair. We have shown that the binding of TRIM72 to the phospholipid membrane induces its self-assembly, which enhances its ubiquitination activity via intermolecular RING domain dimerization. In conclusion, we propose an activation mechanism of higher-order TRIM72 assembly for Ub transfer on the phospholipid membrane.
Results
Crystal structure of dimeric TRIM72
TRIM72 is a member of the class IV TRIM proteins, which is the largest subtype among the 11 groups of TRIM proteins. Its domain organization includes an RBCC domain followed by a PRY motif associated with a SPRY domain identified in splA kinase and ryanodine receptors (PRYSPRY) domain (Fig. 1a)53. TRIM72 is a tight dimer, as determined by size exclusion chromatography (SEC) coupled with multiangle light scattering (SEC–MALS; Extended Data Fig. 1a,b), and its overall shape was determined to be elongated in solution by SEC coupled with small-angle X-ray scattering (SEC–SAXS; Extended Data Fig. 1c–f). To obtain a high-resolution model, numerous constructs were generated, purified and crystallized; however, most of them diffracted poorly. After introducing point mutations and deletions, we were finally able to resolve molecular models of mouse TRIM72, including RING-deleted (ΔRING, 2.75 Å; the highest resolution is in parentheses), full-length (FL, 4.6 Å) and wild-type (WT, 7.1 Å) TRIM72. Details of the constructs and crystal structures used are given in Supplementary Table 1 and Table 1.
a, Domain organization of TRIM72. Each domain and small motif is shown as follows: RING (green); L1 linker, B-box (orange); H1 helix (pink); helix-turn-helix, H2 helix (cyan); L2 linker (mint); H3 helix (yellow); and PRYSPRY (magenta). The residues are numbered based on the mouse TRIM72 sequence. b, Overall structure of mouse TRIM72 represented by a surface model with a ribbon diagram. The color of each domain corresponds to the domain architecture in a. The second protomer is labeled with a prime symbol (′) and is shown in a less-vibrant color. The structural models were derived from mouse TRIM72 ΔRING (2.75 Å), except for the RING domain (green), which came from TRIM72 FL (4.6 Å).
The overall structure of the TRIM72 dimer has an elongated shape (Fig. 1b and Supplementary Fig. 1). The coiled-coil domains (CCDs) form the main scaffold of an antiparallel dimer with a length of approximately 17 nm (Supplementary Fig. 2a–e). In the middle of the CCDs, the structural core consists of the hendecad repeated region of the H1:H1′ helices (prime indicates the second protomer of the dimer) and the H3:H3′ helices followed by a pair of PRYSPRY domains (Supplementary Fig. 2f). Between the H1:H1′ helices and PRYSPRY domains, the H3:H3′ helices are sandwiched to interact with both domains. One side of the H3:H3′ helices is wrapped in a four-helical bundle with the hendecad H1:H1′ helices, and the other side contacts the β-sheet of PRYSPRY through hydrophobic interfaces, hydrogen bonding and ionic interactions (Supplementary Fig. 2g,h). Owing to these interdomain interactions, two PRYSPRY domains are located on the opposite side of the CCDs across the H3:H3′ helices (Fig. 1b).
At the N terminus, a catalytic RING exists in a bent conformation near the other protomer of the CCD (Supplementary Fig. 3a). Next to the RING domain, the B-box domains are located at each end of the CCDs and contact the H1 and H2′ helices of the CCDs in a hydrophobic manner (Supplementary Fig. 3b). The overall structure of TRIM72 obtains a complete model of a member of the TRIM family, which enabled us to investigate the structure–function relationship in detail.
TRIM72 binds to the membrane via its PRYSPRY pair
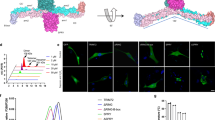
PRYSPRY comprises two antiparallel β-sheets: one sheet (β3, β5–β8 and β12) forms a concave surface, and the other sheet (β1, β2, β4, β9–β11 and β13) forms a convex surface (Fig. 2a)37. The TRIM72 dimer contains two PRYSPRY domains that exhibit unique structural characteristics. First, several arginines (R356, R368, R369, R371 and R386) and lysines (K317, K330, K389, K398, K460 and K462) are situated on the concave surface, consisting of variable loops (VLs), and contribute to its positive charge (Fig. 2a and Extended Data Fig. 2). Second, each PRYSPRY domain displays a concave surface that is positively charged toward the same side, facing opposite the CCDs (Fig. 2b). Finally, the orientation of the PRYSPRY pair is strictly maintained by its interaction with the H3:H3′ helices (Fig. 2b and Supplementary Fig. 2g,h). Through extensive protein–lipid overlay analysis, it was revealed that TRIM72 specifically recognizes negatively charged phospholipids such as PS and phosphatidylinositol but not phosphatidylcholine, phosphatidylethanolamine, oxidized phospholipids or sphingolipids (Extended Data Fig. 3). Flow cytometry analysis showed that TRIM72 also interacted with PS-containing liposomes (PS liposomes) with a much higher affinity for 30 mol% PS than for 10 mol% PS (Fig. 2c). This suggests that clustered and negatively charged phospholipid domains are required for TRIM72–PS binding.
a, Ribbon structure of the PRYSPRY domain of TRIM72. Positively charged lysine and arginine residues are shown in a stick model and labeled. Ct indicates carboxyl terminus. b, Electrostatic potential surfaces of PRYSPRY domains. The positively charged surface of each PRYSPRY domain is maintained by the interaction between H3:H3′ helices (yellow). The second protomer is labeled with a prime symbol (′). c, Saturation curves for TRIM72 binding to PS liposomes (lipo) with PS concentrations of 30 mol% (red) and 10 mol% (blue). d, Liposome coflotation assay with TRIM72 WT and mutants using sucrose density gradients. The fractions collected in tubes are indicated from top to bottom. TRIM72 was detected with an anti-TRIM72 antibody. PC, phosphatidylcholine-containing liposomes; PS, PS liposomes (30 mol% PS). WB, western blot. X and O indicate non-binding and binding, respectively. e, Subcellular fractionation of TRIM72 in C2C12 myoblasts. Membrane (Mem) and cytosol (Cyto) fractions are indicated. The experimental scheme (top) was created with BioRender.com. Western blotting results are shown in Supplementary Fig. 4. Independent experiments were performed in triplicate. Data are presented as mean values with error bars representing s.d. Details of the mutants used in the experiments are provided in Supplementary Table 1.
To investigate the interaction between PRYSPRY domains and negatively charged phospholipid domains, we conducted liposome coflotation assays using the TRIM72 mutants K2D (K460D, K462D) and R3E (R368E, R369E, R371E), which introduce charge reversal in the PRYSPRY domain. In contrast to WT, the charge-reversal mutants did not exhibit any interaction with PS liposomes (Fig. 2d). We also examined the orientations of the PRYSPRY domains with respect to the membrane by generating a TRIM72 mutant, ΔH3 (deletion of residues 272–281), that disrupted the orientation of the PRYSPRY domains. As expected, we observed a loss of interaction between PS liposomes and the deletion mutant, confirming the importance of the orientation of PRYSPRY domains in recognizing the negatively charged phospholipid membrane (Fig. 2d). Next, we measured the binding affinity between TRIM72 and PS liposomes. The binding constant KD was calculated to be 9.9 nM, which is similar to the affinity between annexin V and PS (Extended Data Fig. 4a,b)54. We observed that dimeric glutathione S-transferase (GST)-fused H3–PRYSPRY interacted with PS liposomes with a KD of 76.7 nM, which was 7.7-fold lower than that of WT (Extended Data Fig. 4a,g). However, monomeric maltose-binding protein (MBP)-fused H3–PRYSPRY did not bind to PS liposomes, similar to the defective mutants RBCC, K2D, R3E and ΔH3 (Extended Data Fig. 4). Thus, we interpret this result to mean that avidity is critical for PS recognition by TRIM72.
The results were also confirmed in vivo upon fractionation of both the membrane and cytosol in C2C12 myoblast cells after transfection to express human TRIM72. In contrast to TRIM72 WT, the mutants (K2D, R3E and ΔH3) exhibited significant defects in membrane localization (Fig. 2e and Supplementary Fig. 4). Therefore, under physiological conditions, TRIM72 binds to the lipid membrane using its positively charged PRYSPRY domains. In conclusion, our results demonstrate the importance of the orientation, avidity and charge distribution of the PRYSPRY domains in recognizing negatively charged phospholipid domains.
TRIM72 forms a higher-order assembly on the membrane
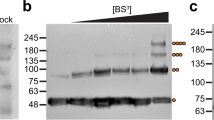
TRIM72 is known to oligomerize in an oxidation-dependent manner and is critical for vesicle nucleation to facilitate rapid membrane repair18. Under oxidizing conditions by the addition of H2O2, oligomerized TRIM72 was observed only in PS liposomes (Supplementary Fig. 5a). This result indicates that TRIM72 self-oligomerizes on the negatively charged phospholipid membrane. To further characterize the multimerization of TRIM72, we treated TRIM72 with various sulfhydryl- and amine-reactive cross-linkers in the presence or absence of PS liposomes (Supplementary Fig. 5b). Higher-order TRIM72 oligomers were observed under conditions in which they were bound to PS liposomes. Time-course cross-linking analysis showed that TRIM72 WT gradually oligomerized over time, but the mutants (K2D, R3E and ΔH3) produced fewer higher-order oligomers because they were unable to bind negatively charged PS liposomes (Extended Data Fig. 5). These findings suggest that TRIM72 binds to the negatively charged phospholipid membrane and then oligomerizes into higher-order assemblies.
Interestingly, human TRIM72 purified from mammalian cells cross-linked much faster than mouse TRIM72 from bacterial cells, likely due to its prior binding to endogenous membranes in mammalian cells (Extended Data Fig. 6a). In fact, during affinity chromatography (AC) and SEC, mammalian-expressed human TRIM72 was copurified with microvesicles that were approximately 100 nm in diameter (Extended Data Fig. 6b,c). These microvesicles were identified as originating from extracellular vesicles or junctional membranes and were enriched with TRIM72 and Na+–K+ ATPase (Supplementary Data 1). When microvesicles were visualized by transmission electron microscopy (TEM) after negative staining, long stripe patterns were observed on the microvesicles (Extended Data Fig. 6c). To confirm whether the patterns originated from oligomerized TRIM72, we reconstituted TRIM72 proteoliposomes in vitro and characterized them. TRIM72 proteoliposomes were successfully isolated by SEC after incubation with TRIM72 and small PS liposomes approximately 100 nm in diameter and showed an SEC profile similar to that of TRIM72-bound microvesicles from mammalian cells (Extended Data Fig. 6b). In the negative-staining TEM images, long stripe patterns were also observed on the reconstituted proteoliposomes, which were similar to those found on microvesicles (Extended Data Fig. 6d).
To investigate how TRIM72 oligomerizes on the phospholipid membrane, we analyzed TRIM72 proteoliposomes using cryogenic electron tomography (cryo-ET) (Extended Data Fig. 6e and Supplementary Videos 1 and 2). We observed higher-order TRIM72 assembly on both surfaces of the lipid bilayer, with the inner surface being better resolved than the outer surface. The TRIM72 assembly exhibited vertical column-like densities standing on the lipid membrane at regular intervals and a unique bridge-like structure connecting these column densities horizontally.
Molecular model of the higher-order TRIM72 assembly
To obtain a detailed model of TRIM72 assembly on the phospholipid membrane, we averaged subtomograms containing TRIM72-repeating units from tomograms of TRIM72 proteoliposomes. This allowed us to generate a density map of the higher-order TRIM72 assembly complexed with the lipid bilayer at an overall resolution of 25 Å (Fig. 3a,b, Extended Data Fig. 7 and Table 2). The TRIM72 assembly appeared in a regular pattern with 6-nm spacing on the lipid bilayer, composed of vertical column densities and a planar density spanning the region between them (Fig. 3c and Extended Data Fig. 8c). Double column densities connected the planar density above the lipid bilayer, forming the characteristic bridge-like structure of the TRIM72 assembly. This structure was observed in detail from the view perpendicular to the lipid surface (Fig. 3b,c).
a, Cryo-ET maps of the reconstituted TRIM72 proteoliposome. Top view (left) and front view (right) are shown with subtomogram averaging reconstruction density contoured at 1 σ. b, Higher-order TRIM72-assembly model on the lipid bilayer. Top left, front view (perpendicular to the lipid surface) of the TRIM72-assembly model with a subtomogram averaging map contoured at 1 σ. Top right, the black box in the front view is enlarged to a close-up view. The top view (parallel to the lipid surface, bottom left) and a sliced view (bottom right) of the TRIM72 assembly. The black boxes in the top view are represented as ribbon diagrams in d,e. The highlighted TRIM72 protomers in the dimer are colored red and blue, respectively. The lipid bilayer models are shown in stick representation in green (top right). Subtomogram averaging reconstruction maps of the TRIM72 assembly and lipid bilayer are colored in white and pink, respectively. c, Assembly pattern of TRIM72 as observed in subtomogram averaging reconstruction. Averages of z slices correspond to the middle and front view in b. The relative intensities according to the y axis (right; red) and the x axis (bottom; blue) were calculated from the averaged z slices of the front view. Numbers in italics (1–5) indicate each column-like density corresponding to the PRYSPRY domain. Note that the regular intervals between the PRYSPRY densities are 6 nm (bottom). Black scale bars, 5 nm in a–c. d, Interface between B-boxes. Interacting residues and zinc ions are shown in stick and sphere models, respectively. e, Interface between H1 helices in the CCDs. f, Circle plots of cross-linked cysteine residues of TRIM72 with or without PS liposomes. Frequently identified cross-linked pairs are shown as thick lines. Homotypic cysteine-bridged pairs are shown in red and were identified in the presence of PS liposomes. g, Liposome co-sedimentation assay of TRIM72 WT and mutants. The experimental scheme is shown at the top; the image was created with BioRender.com. PC liposomes, phosphatidylcholine-containing liposomes; pel, pellet; sup, supernatant.
Using X-ray crystallography and subtomogram averaging, we investigated the molecular architecture of the higher-order TRIM72 assembly complexed with the phospholipid bilayer. We were able to fit approximately 18 dimers of TRIM72 as a rigid body on the phospholipid bilayer with an area of 1,400 nm2 approximately with the guidance of crystal packing (Fig. 3b). The PRYSPRY domains are located on the double column-like density, while other domains, such as CCDs, B-boxes and RINGs, occupy the planar density above the lipid bilayer. The assembly allows for the positively charged surfaces of the PRYSPRY domains to be in close proximity to the surface of the phospholipid bilayer (Fig. 3b).
There are two interactions between each dimer in the TRIM assembly. The major interaction is between B-boxes, which mediate dimer-of-dimer formation through Gly106 and Ser110′ interacting with each other by hydrogen bonding (Fig. 3b,d and Supplementary Fig. 6a,b). The interaction was confirmed by cross-linking mass spectrometry (CLMS), in which cross-linked residues between the B-boxes (Cys105–(cross-linker)–Cys105′ and Cys105–(cross-linker)–Cys108′) were detected only in the presence of PS liposomes (Fig. 3f and Supplementary Fig. 7a). Mutations at the B-box interface (TRIM72G106R and TRIM72S110R) resulted in defective membrane binding and assembly (Fig. 3g).
CCD-mediated interactions also play a crucial role in ensuring that the PRYSPRY domains are aligned in the same orientation (Fig. 3b,e). These CCD interactions are organized by a hydrophobic interface mediated by Met138, which is exposed on the outer side of the H1 helix in the CCD. Mutations at the Met138 site (TRIM72M138A) resulted in a severe defect in higher-order TRIM72 assembly on PS liposomes, despite maintaining membrane binding (Extended Data Fig. 8a,b). However, the TRIM72M138R variant had a higher binding affinity to PS liposomes and clearer patterns of TRIM72 assembly than TRIM72 WT (Extended Data Fig. 8c and Supplementary Videos 3 and 4). Subtomogram averaging of TRIM72M138R revealed that the density map of the TRIM72M138R assembly was similar to that of TRIM72 WT (Extended Data Fig. 8d–g). In addition, cross-linked residues (Cys144–(cross-linker)–Cys144′) were detected near the interface between CCDs, supporting the observation of proximity between CCDs (Fig. 3f and Supplementary Fig. 7a).
The distance between cross-linked cysteines identified by CLMS was used to validate the model of higher-order TRIM72 assembly. The TRIM72-assembly model showed minimized distances between cross-linked cysteines, whereas the dimer or dimer-of-dimer models showed outliers above 50 Å (Supplementary Fig. 7b and Supplementary Data 2). In conclusion, by integrating structural and biochemical analyses, we determined the molecular model of the higher-order TRIM72 assembly complexed with the negatively charged phospholipid bilayer. The TRIM72 assembly aligns the PRYSPRY domains on the surface of the phospholipid membrane. Furthermore, intermolecular domain interactions cooperatively affect TRIM72 assembly to maintain tighter binding with the negatively charged phospholipid membrane.
Ubiquitination activity of TRIM72 is enhanced on the membrane
Screening of the E2 conjugating enzyme revealed that TRIM72 exhibits ubiquitination activity with ubiquitin-conjugating enzyme E2 D (UBE2D) enzymes (Supplementary Fig. 8a). However, TRIM72 expressed in bacterial cells, even with duplicated RING domains (2×RING), did not show detectable ubiquitination activity (Supplementary Fig. 8b), in contrast to TRIM72 expressed in mammalian cells (Supplementary Fig. 8a). We also checked the phosphorylation status of these TRIM72 proteins but found no difference between them (Supplementary Fig. 8c). Hence, we investigated whether the ubiquitination activity of TRIM72 could be enhanced by phospholipid membrane binding. We reconstituted TRIM72 proteoliposomes and evaluated their ubiquitination activity. Indeed, ubiquitination activity increased only in reconstituted TRIM72 proteoliposomes, not in TRIM72 alone or liposomes alone (Fig. 4a). This indicates that the ubiquitination activity of TRIM72 is suppressed in solution. To investigate how ubiquitination activity is suppressed, we focused on the residue next to the last zinc-coordinating cysteine, called the ‘linchpin’. The linchpin is critical to Ub transfer because it forms hydrogen bonds with both the main-chain carbonyl oxygen atoms of E2 enzymes (Gln92 in UBE2Ds and Lys94 in UBE2N) and Ub (Arg72)55,56. While the canonical linchpin is an arginine residue, the corresponding residue in TRIM72 is a suboptimal glutamine residue (Gln57). We mutated the glutamine linchpin to arginine (Q57R) and then tested the ubiquitination activity. The TRIM72Q57R mutant exhibited much higher ubiquitination activity than the TRIM72-WT protein (Fig. 4b). The membrane-bound TRIM72Q57R mutant also showed much higher ubiquitination activity than the soluble TRIM72Q57R mutant, although soluble TRIM72Q57R exhibited partially restored ubiquitination activity. These results clearly demonstrate that the ubiquitination activity of TRIM72 is suppressed in the soluble state due to the suboptimal linchpin. Similar to reconstituted proteoliposomes, the mammalian-expressed TRIM72Q57R mutant possessed highly enhanced ubiquitination activity in both fractions containing vesicle-bound and vesicle-free soluble forms (Supplementary Fig. 8d).
a, Ubiquitination activity of TRIM72 in the presence or absence of PS liposomes. b, Increased ubiquitination activity of TRIM72Q57R in the presence or absence of PS liposomes. c, Ubiquitination activity of TRIM72Q57R and TRIM72Q57R/L74R in the presence or absence of PS liposomes. d, Ubiquitination activity of endogenous microvesicle-bound or free-solution states of TRIM72Q57R and TRIM72Q57R/L74R. e, Ubiquitination activity of RING and 2×RING constructs with TRIM72 WT and TRIM72Q57R. f, Ubiquitination activity of RING and 2×RING constructs with TRIM72Q57R and TRIM5α. TRIM72 proteoliposomes were reconstituted and further separated by SEC (a–c). TRIM72 microvesicles were purified from HEK293T cells and isolated by SEC (d). Poly-Ub is represented as a polyubiquitination ladder. Bands close to 140 kDa were assumed to be ubiquitin-like modifier activating enzyme 1 (UBA1)~Ub. Ub was detected with an anti-Ub antibody. Strep-tagged TRIM72 or intact TRIM72 was detected using an anti-Strep-tag antibody or an anti-TRIM72 antibody, respectively (a–d). The superfolder variant of green fluorescent protein (sfGFP)-fused RING and 2×RING constructs were detected with an anti-GFP antibody (e,f). Phospholipids were stained with Sudan Black B (a–c). Microvesicles were detected with an anti-Na+–K+ ATPase α1 antibody (d). Independent experiments were performed in triplicate. TRIM72 WT, green; TRIM72Q57R, pink; TRIM72Q57R/L74R, cyan; TRIM72 RING_WT, yellow; TRIM72 RING_Q57R, light pink; TRIM5α RING, magenta.
Ub transfer is catalyzed by transient RING dimerization
In our crystal structures of TRIM72, the key residue Gln57 in the RING domain was observed near Arg207′ in the CCD through the bent RING conformation (Supplementary Fig. 3a). However, when we superimposed the structure with the TRIM25 RING–UBE2D2–Ub ternary complex43, we found that the bent RING conformation was not accessible to E2–Ub intermediates due to steric hindrance with the CCDs. To further explore the conformation of the RING domain, we used the SAXS refinement through flexibility (SREFLEX) approach (Supplementary Fig. 9a–c; statistics of the SAXS analysis are summarized in Supplementary Table 2)57. Based on the experimental SAXS profiles, the calculated models showed that the RING domain is highly dynamic and does not prefer the bent conformation (Supplementary Fig. 9a). We also observed that the electron density map of the RING domain appears in only one of the four crystal structures of TRIM72 FL, suggesting that the RING domain is intrinsically flexible. Additionally, an AlphaFold58 prediction suggested that TRIM72 has the extended conformation of the RING domain, similar to one of the models calculated by SREFLEX. Not only structural evidence but also biochemical data indicated that there was no difference in ubiquitination activities between the TRIM72Q57R and TRIM72Q57R/R207E mutants (Supplementary Fig. 9d). Therefore, we conclude that the RING domain is highly flexible in both the TRIM72 dimer and oligomer on the phospholipid membrane.
Next, we investigated whether dimerization of the RING domain is necessary for Ub transfer. In contrast to the TRIM72 RING domains in the dimer model, which are approximately 20 nm apart, the RING domains are located close to each other in the higher-order assembly model on the membrane (Fig. 3b). This suggests that TRIM72 RING domains transiently dimerize between dimeric TRIM72 and another dimeric TRIM72 in the oligomeric assembly. To determine whether dimerization of the RING domains is essential for Ub transfer, we introduced a mutation to disrupt the dimeric interface on the RING domain. Structural comparison with the TRIM25 RING dimer complexed with UBE2D2–Ub43 predicted that Leu74 of TRIM72 (corresponding to Val72 in TRIM25) is critical to the dimerization of the RING domain. Thus, we replaced Leu74 of TRIM72 (L74R) with a bulky and hydrophilic arginine residue to block the dimeric interface. Because the WT activity was weak, we compared the ubiquitination activity of TRIM72L74R with that of the TRIM72Q57R-mutant background. In contrast to the highly active TRIM72Q57R mutant, TRIM72Q57R/L74R completely lacked activity in both membrane-bound and soluble forms (Fig. 4c). Furthermore, we observed similar results not only for the reconstituted TRIM72 proteoliposomes but also for mammalian-expressed TRIM72 bound to endogenous microvesicles (Fig. 4d), suggesting that TRIM72 RING domains dimerize and activate in the higher-order assembly on the membrane. Finally, we dissected the dimerization effect of the RING domain without other domain interactions of TRIM72 by carrying out the ubiquitination assay using TRIM72 RING and 2×RING based on the TRIM72Q57R background. In contrast to the low ubiquitination activities of TRIM72 RING and 2×RING, as previously shown, TRIM72 2×RING_Q57R showed significantly increased activity compared to that of monomeric TRIM72 RING_Q57R (Fig. 4e). This finding is similar to the dimeric activation mode of TRIM5α RING, although the activity of TRIM72 2×RING_Q57R was still lower than that of TRIM5α RING (Fig. 4f). These results suggest that RING dimerization is necessary to activate the mechanism of TRIM E3 Ub ligase. Therefore, we conclude that the intermolecular RING domains can transiently dimerize in the higher-order TRIM72 assembly on the phospholipid membrane for Ub transfer.
TRIM72 assembles on the membrane to overcome Ca2+ influx
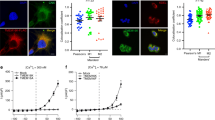
In addition to its ubiquitination activity, we investigated the effect of Ca2+ on the interaction between TRIM72 and PS. Protein–lipid overlay analysis revealed that Ca2+ inhibited the interaction between TRIM72 and PS (Fig. 5a and Extended Data Fig. 3). However, the interaction was completely restored by ethylene-bis(oxyethylenenitrilo)tetraacetic acid (EGTA), a Ca2+-chelating agent (Fig. 5a), indicating that Ca2+ directly inhibits the interaction between TRIM72 and PS. Our structural and biochemical evidence suggests that the positively charged PRYSPRY domains of TRIM72 and Ca2+ compete with the negatively charged head group of PS via ionic interactions. This finding partially conflicts with a previous report that TRIM72 facilitates vesicle delivery to the damaged site in a Ca2+-independent manner18. We carefully assessed the inhibitory effect of Ca2+ by adding each factor one by one. Real-time flow cytometry analysis demonstrated that TRIM72 could not bind to PS liposomes in the presence of Ca2+ (Fig. 5b,c). Surprisingly, preloaded TRIM72 barely dissociated from PS liposomes upon addition of Ca2+. By contrast, in annexin, a negatively charged loop engages in a Ca2+-mediated interaction with the head group of PS59. This indicates that oligomerization acts as a secondary mode of interaction, strengthening the interaction between TRIM72 and PS-rich domains on the phospholipid membrane. In conclusion, higher-order TRIM72 assembly on the membrane surface, rather than the dimeric form in solution, can overcome Ca2+ influx and is recruited, similar to repair vesicles, to damaged sites of the plasma membrane for wound healing.
a, Protein–lipid overlay analysis showing the interference. TRIM72 proteins were incubated with lipid strips in the presence or absence of Ca2+ and/or EGTA and then probed with an anti-TRIM72 antibody. PE, phosphatidylethanolamine. N/A indicates phospholipid is not applied. b, Flow cytometry analysis comparing the effects of Ca2+ on TRIM72 and annexin V (ANXV). PS liposomes were preloaded with Ca2+ or loaded after Ca2+ exposure and then incubated with TRIM72 or annexin V. Binding was detected using fluorescently labeled sfGFP–TRIM72 and fluorescein isothiocyanate (FITC)–annexin V. Ex488/Em533 nm, excitation at 488 nm/emission at 533 nm. c, Quantification of the data in b. Experiments were performed in triplicate. Data are presented as mean values ± s.d. The PS concentration was 30 mol% in PS liposomes. Ca2+ ion, yellow; TRIM72, green; annexin V, pink.
Discussion
In this study, we demonstrate the assembly of the RING-type E3 ligase TRIM72 into functional oligomers on phospholipid membranes (Fig. 6). Our findings showed that the ubiquitination activity of TRIM72 is suboptimal in solution, and the active conformation is induced by higher-order assembly on the membranes. This event is mediated by the interaction between the negatively charged phospholipid membranes supplied by PS in cells and the positively charged surfaces of PRYSPRY domains oriented in the same direction. As a result, dimeric TRIM72 aligns on the membrane and further self-oligomerizes into a higher-order assembly. The conformation of the higher-order TRIM72 assembly allows RING domains to transiently homodimerize. Finally, E2~Ub intermediates (the tilde denotes a covalent thioester bond) can be recruited into dimerized RINGs of the higher-order TRIM72 assembly on the membrane for efficient Ub transfer.
In the solution state (top), TRIM72 (rose gold) cannot recruit E2~Ub (cyan, yellow) conjugates for Ub (yellow) transfer. The flexibility of the RING domain (pink) at both ends of the dimeric TRIM72 and dynamic Ub-conjugating E2 enzymes is indicated by curved dashed lines with arrows. Although B-boxes and CCDs at both ends are also moved in the perpendicular direction, this is not depicted in this model for clarity. The amino acid frequency of the linchpin is shown in a black-lined box. The linchpin, positioned at the residue following the final Zn2+-coordinated cysteine, was analyzed in 303 sequences of human RING domains (Supplementary Data 3). Note that ~50% of RING domains have a suboptimal linchpin, including TRIM72 (glutamine linchpin). The higher-order TRIM72 assembly on the negatively charged membrane (middle) and the active-state model with E2~Ub conjugates (bottom) are shown. When dimeric TRIM72 is targeted to PS-containing vesicles or PS-enriched plasma membranes, it primarily binds to the membrane via the PRYSPRY domain. Next, higher-order TRIM72 assembly is mediated by cooperative interactions among B-box–B-box and CCD–CCD. In this assembly, dynamic RING domains reduce the motion, and intermolecular RING dimer formation occurs. The next layered TRIM72 oligomer in the assembly is shown in a less-vibrant color. Finally, the functional ternary complex is stable enough for efficient Ub transfer. The model was created with BioRender.com.
There are two reasons why the TRIM72 dimer is unsuitable for Ub transfer with UBE2D~Ub intermediates in solution. First, the RING domain possesses a suboptimal linchpin, glutamine in TRIM72 (Gln57). In contrast to the optimal linchpin, which is conserved as arginine in half of RING-type Ub ligases55 and mutated in human cancers (Fig. 6 and Supplementary Data 3)60,61, the suboptimal linchpin is insufficient as a hydrogen bonding donor to stimulate flexible UBE2D~Ub or UBE2N~Ub intermediates into the closed conformation for robust Ub transfer56,62. We showed that changing the linchpin to the optimal arginine significantly enhanced the ubiquitination activity of TRIM72 both on the membrane and in solution. This finding indicates that the RING domain of TRIM72, due to the suboptimal linchpin, is insufficient to bind a closed conformation of the UBE2D~Ub intermediate. To obtain appropriate binding affinity with E2~Ub, it is essential to dimerize RINGs in known TRIM proteins43,49,63,64. However, the second reason is that RINGs are separated by antiparallel CCDs and laid in monomeric conformations43,49,63,64. Considering the structural prediction obtained with ColabFold65 and experimental evidence supporting the RING dimer, most of the 60 TRIMs (except TRIM15, TRIM40, TRIM45 and class VI TRIMs) have the potential to possess dimerized RINGs in a similar manner (Supplementary Data 4). Therefore, in contrast to the relatively diverse linchpin mechanism, RING dimerization is likely a more general mechanism for the active conformation in the TRIM family43,63,64,66.
We highlight the active conformation induced by RING dimerization in higher-order TRIM72 assembly on the membrane. The TRIM72 assembly is cooperatively derived from the intermolecular B-box interaction and the CCD interaction, leading to close positioning of the RING domains to each other and further dimerization despite very weak affinity in solution. Compared to the higher-order assembly of primate TRIM5α, which binds and restricts human immunodeficiency virus capsids67,68, B-box-mediated interactions are found in the assembly process of both TRIMs. Furthermore, the proximity required to induce RING dimerization is achieved through the interaction between the B-boxes in both TRIMs. Thus, B-box-mediated assembly for functional activation in TRIMs might be a common process. On the other hand, in contrast to the hexagonal and pentagonal assembly of TRIM5α, the bridge-like pattern of TRIM72 assembly is mediated by the interaction between the solvent-exposed hydrophobic surfaces of CCDs, indicating that the structural features of TRIM assembly might vary depending on the target molecules and their interactions. In addition to TRIM72 and TRIM5α, a subset of class IV TRIMs, including TRIM65, potentially oligomerize on flexible double-stranded RNA (dsRNA) filaments through avidity-driven binding between their PRYSPRY domains and the RNA helicase domains of cognate retinoic acid-inducible gene I (RIG-I)-like receptors50. Recent cryo-electron microscopy (EM) analysis of TRIM72 and the crystal structure of TRIM72 ΔRING also revealed that the CCD ends are dynamic69,70. These results suggest that the flexibility of long TRIM CCDs is suitable for recognizing curved cellular structures, such as phospholipid membranes and viral capsids, and filamentous structures, such as dsRNA and DNA. The current findings provide a conceptual basis for the study of TRIM proteins. Compared to other TRIM PRYSPRY domains, which have been well studied for their interactions with their binding molecules36,50,51,68, these PRYSPRY domains use the concave surface52 to recognize their interacting molecules (Extended Data Fig. 2a–f). The crucial residues for binding are located on VL1, VL3–β6–VL4 and VL6 (Extended Data Fig. 2g), which suggests that substrate specificity is determined by sequence diversity around the VL regions. In contrast to other PRYSPRY domains, TRIM72 PRYSPRY domains show more distributed positively charged residues on the VL regions, which contributes to their unique basic properties for membrane binding (Fig. 2a,b and Extended Data Fig. 2a).
TRIM72’s interaction with the membrane involves spatially controlled ubiquitination activity in muscle differentiation and membrane-repair processes. Our findings suggest that higher-order TRIM72 assembly is essential for its membrane-repair function, as previously reported in muscle injury18. We have elucidated the molecular basis of TRIM72 oligomerization, including the preference for Ca2+-independent PS binding, key residues for oligomerization, the architecture of the higher-order TRIM72 assembly and activation of E3 ligase activity. In the context of membrane repair, the ESCRT machinery plays a vital role, which is closely related to Ub signaling3,8,9,14. Based on our observations, we speculate that self-ubiquitination or the ubiquitination of target substrates may contribute to the membrane-repair process3. However, further investigation is needed to understand the links between ubiquitination and membrane-repair mechanisms. Although some features are unique to TRIM family proteins or only a subset of them, the extensive information gained from the current structural and biochemical study of TRIM72 offers broad insights into RING-type E3 Ub ligases.
Methods
Cloning, protein expression and purification
FL mouse Trim72 and human TRIM72 genes were amplified by PCR and cloned into bacterial (modified pMAL and pRSF) and mammalian (modified pHM6) expression vectors, respectively. The domain-deletion and point mutant constructs were cloned by PCR-based mutagenesis. All constructs (Supplementary Table 1) were verified by DNA sequencing. WT TRIM72 and mutants with an N-terminal His6–MBP tag were expressed in Escherichia coli BL21(DE3) cells and purified. For flow cytometry analysis, mouse TRIM72 tagged with N-terminal His6–sfGFP71 was expressed and purified. Purification of GST–H3–PRYSPRY was carried out as previously described37. Cells were induced at an OD600 of 0.7 with 500 μM isopropyl β-d-1-thiogalactopyranoside and grown with 200 μM ZnCl2 (depending on the presence of DNA encoding the RING and B2-box domains) at 18 °C for 18 h. Proteins were purified by AC followed by ion exchange (IEX) and SEC with equilibrated SEC buffer (50 mM Tris-HCl, pH 8.0, 300 mM NaCl and 1 mM Tris(2-carboxyethyl)phosphine (TCEP)). The N-terminal MBP tag was cleaved by on-column tobacco etch virus protease digestion. The cleaved MBP tag and tobacco etch virus protease were removed by AC and IEX. The quality of TRIM72 WT and the mutant proteins was verified by circular dichroism and thermal shift assays. Human TRIM72 WT and mutants with an N-terminal 2× Strep tag (MASAWSHPQFEKGGGSAWSHPQFEKGS) were expressed in HEK293T mammalian cells after transfection with polyethyleneimine. After 48 h, the cells were washed with Dulbecco’s phosphate-buffered saline (DPBS) and resuspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl and 1 mM TCEP). After lysis with an ultrasonic probe and centrifugation (27,216g, 4 °C for 1 h), the supernatants were loaded onto a StrepTrap HP column (Cytiva, 28907548). The proteins were eluted with Strep elution buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM TCEP and 2.5 mM d-desthiobiotin) and then subjected to SEC using Superose 6 Increase 10/300 GL columns (Cytiva, 29091596). Each fraction containing TRIM72 was subjected to a ubiquitination assay and EM. Rhesus TRIM5α RING and tandem repeats of RING constructs were purified using His6–sfGFP tags followed by IEX. Plasmids encoding UBA1 (human UBA1, Addgene, 34965)72 and UBE2D3 (human UBE2D3, Addgene, 15784)73 were obtained from Addgene (http://addgene.org). E1 and E2 enzymes were purified with general bacterial expression protocols. Ub was prepared as described in previous studies74.
Crystallization and data collection
Mouse TRIM72 proteins (5–10 mg ml−1) were crystallized by the vapor-diffusion method with a 1:1 mixture of crystallization reagents and proteins. Although many crystals were obtained from the initial crystallization screen at 22 °C, most of these crystals were brittle and poorly diffracted to a resolution of only 7 Å. To improve crystal quality and change crystal packing, we designed numerous constructs by modifying the residues suspected to be exposed on the surface. Among them, distinct crystal forms were obtained by crystallizing a construct containing five point mutations and omitting the N-terminal six amino acids and the C-terminal seven amino acids (Supplementary Table 1). Indeed, the primitive monoclinic crystals showed greatly improved diffraction patterns at resolutions of up to 3.5 Å. After further optimization by limited proteolysis with elastase, mouse TRIM72 ΔRING crystals grown at 4 °C diffracted up to 2.75 Å. The crystallization conditions were as follows: 0.8 M NaCl and 8% (vol/vol) ethanol (WT, 7.1 Å; constructs, highest resolution); 100 mM MES-NaOH, pH 6.5, 200 mM MgCl2, 0.5% (wt/vol) polyvinylpyrrolidone K 15 and 30% (wt/vol) pentaerythritol propoxylate (5/4 PO/OH) with microseeding (FL, 4.6 Å); 100 mM HEPES-NaOH, pH 7.5, 230 mM MgCl2, 100 mM CsCl and 34.16% (wt/vol) pentaerythritol ethoxylate (3/4 EO/OH) with Al’s oil (FL, 3.5 Å); 100 mM HEPES-NaOH, pH 7.5, 250 mM MgCl2 and 37% (wt/vol) 5/4 PO/OH (FL/C242S, 5.2 Å); 100 mM Tris-HCl, pH 8.5 and 30% (vol/vol) PEG 400 with microseeding (ΔRING, 2.75 Å); and 100 mM Tris-HCl, pH 8.5 and 8% (vol/vol) PEG 8000 (ΔRING, 3.3 Å). Crystals were flash frozen in liquid nitrogen under cryoprotective conditions. Datasets were collected at the following synchrotron facilities: the Pohang Accelerator Laboratory in South Korea and the Photon Factory and Super Photon Ring-8 GeV (SPring-8) in Japan. Data processing, integration and scaling were carried out with DENZO and SCALEPACK from the HKL-2000 suite75. Detailed statistics are summarized in Table 1.
Structure determination and refinement
The phases of mouse TRIM72 ΔRING were solved by molecular replacement using phenix.phaser76 with human TRIM72 PRYSPRY (PDB 3KB5) as a search model37. The phases were improved by combining single-wavelength anomalous dispersion from two zinc ions in the B-box domain. An unassigned model of the B2-box domain and the CCD was manually built with Coot77 and rebuilt with phenix.autobuild between refinement cycles78. The phases of the other crystals were solved by molecular replacement with a monomer of ΔRING as a search model. Detailed refinement statistics are summarized in Table 1. Among the FL mouse TRIM72 structures, an electron density map corresponding to the RING domain was found only in the trigonal crystals of FL and diffracted up to a resolution of 4.6 Å. In the other crystal systems, we could not find sufficient electron density for the RING domain; accordingly, we believe that the RING domain does not participate in crystal contacts in these crystal systems and that it is also very dynamic. The electron density map of the RING domain was found at the outer surface of another protomer’s CCD and was separated by two bulky difference density maps. Two zinc ions were placed at the center of each separated difference map. Interestingly, the distance between the zinc ions was approximately 14 Å, the interzinc distance in the classical C3HC4-type RING domain79. Due to the low resolution, we initially built the RING domain coordinates by homology modeling using SWISS-MODEL80 and then fitted the model into residual difference maps and refined it with phenix.refine81. The quality of the final model was validated by MolProbity82.
Structural and multiple-sequence analyses
Structural analysis was performed using the PISA server in the CCP4 suite83. Structure-based sequence alignments were conducted with PROMALS3D84. Sequences were aligned with Clustal Omega85 and further analyzed with BioEdit software86. The logo graph was generated with WebLogo (https://weblogo.berkeley.edu/logo.cgi). Structural figures were prepared with PyMOL (Schrödinger) and UCSF ChimeraX87.
Small-angle X-ray scattering
Purified mouse TRIM72-WT and ΔRING proteins were prepared in SAXS buffer (25 mM Tris-HCl, pH 8.0, 300 mM NaCl, 1 mM TCEP and 1 mM dithiothreitol (DTT)). SAXS data were collected at synchrotron beamlines 4C at the Pohang Accelerator Laboratory in South Korea and 10C at the Photon Factory in Japan. For SEC–SAXS experiments, proteins were loaded onto Superdex 200 Increase 10/300 GL resin (Cytiva, 28990944) equilibrated with SAXS buffer, after which datasets were collected from peak fractions and normalized buffer fractions for background subtraction. Data processing was performed using the ATSAS software package88. For flexible refinement of the TRIM72-WT model, two RING domains of dimeric TRIM72 FL were defined as individual rigid bodies and fitted to experimental curves for mouse TRIM72 WT using SREFLEX software in ATSAS online57. The final models were superimposed on 25 refined models. Details of data collection and parameters are summarized in Supplementary Table 2.
Size exclusion chromatography with multiangle laser light scattering
SEC–MALS was performed using an ÄKTA FPLC system (Cytiva) coupled with a Wyatt miniDAWN TREOS detector (Wyatt Technology). Protein was loaded on a Superdex 200 Increase 10/300 GL column equilibrated with MALS buffer (50 mM Tris-HCl, pH 8.0, 300 mM NaCl and 2 mM TCEP). Ovalbumin (Sigma-Aldrich, A7641) was used as an isotropic scatterer for calibration and normalization of the detectors. Light scattering was measured and analyzed using ASTRA V software (Wyatt Technology).
In vitro ubiquitination assay
An in vitro E2 screening assay was performed using 50 nM UBA1, 500 nM of each E2 enzyme in a set (Boston Biochem, K-980B) and 10 μM Ub with 2 μM mouse TRIM72 constructs in ubiquitination buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl and 1 mM TCEP) at 37 °C for 1 h. In vitro ubiquitination reactions were performed with 50 nM UBA1, 2 μM UBE2D3 and 3 μM Ub with 1 μM mouse TRIM72, rhesus TRIM5α or 0.2 μM human TRIM72. In the ubiquitination assay with TRIM72 mutants based on the Q57R background, 0.5 μM UBE2D3 was used instead of 2 μM UBE2D3 to assess the mutation effects of TRIM72. All reactions were initiated by the addition of 5 mM ATP and MgCl2. The reaction was stopped by the addition of sodium dodecyl sulfate (SDS) sample buffer containing β-mercaptoethanol (β-ME). The samples were boiled at 95 °C for 10 min and then resolved by SDS‒PAGE for further analysis. The polyubiquitinated chains were detected by western blot analysis using the Ub-specific antibody P4D1 (Santa Cruz, sc-8017) or VU-1 (LifeSensors, VU101), while TRIM72 WT and mutants were detected using a rabbit polyclonal anti-TRIM72 (residues 90–277) antibody19, and sfGFP-fused TRIM constructs were detected using a mouse monoclonal anti-GFP antibody (Santa Cruz, sc-9996). Phospholipids were stained with Sudan Black B (Sigma-Aldrich, 199664). TRIM72-bound microvesicles were detected with a mouse monoclonal anti-Na+–K+ ATPase α1 antibody (Santa Cruz, sc-21712).
Protein–lipid overlay assay
The concept of the protein–lipid overlay assay has been described previously89. The following lipid strips were used to screen the lipid specificity of TRIM72: PIP Strips (Thermo Fisher Scientific, P23750), Inositol Snoopers (Sigma-Aldrich, 330500), Oxidized Phospholipid Snoopers (Sigma-Aldrich, 330501) and Sphingolipid Snoopers (Sigma-Aldrich, 330503). The lipid strips were blocked with 3% (wt/vol) bovine serum albumin (BSA; A0100, GenDEPOT) in TBS-T (10 mM Tris-HCl, pH 7.4, 140 mM NaCl and 0.1% (vol/vol) Tween-20) at room temperature (22 °C) for 1 h. After blocking, the lipid strips were washed five times with TBS-T and further incubated with 1 μg ml−1 mouse TRIM WT, 3% (wt/vol) BSA and TBS-T at 4 °C overnight. To determine the effects of Ca2+, 2.5 mM CaCl2 and 10 mM EGTA (Sigma-Aldrich, E3889) were used as Ca2+-chelating agents. Lipid binding specificity was detected by using a rabbit polyclonal anti-TRIM72 antibody19.
Liposome preparation
Liposomes were freshly prepared by vortexing and extrusion methods. The compounds 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC; Sigma-Aldrich, 850375C), 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS; Sigma-Aldrich, 840035C), 1,2-dipalmitoyl-sn-glycero-3-phospho-l-serine (DPPS; Sigma-Aldrich, 840037P) and cholesterol (Sigma-Aldrich, C8667) were dissolved in chloroform or a chloroform–methanol (2:1) solution. One micromole of lipids with the indicated compositions was evaporated in a glass vial under a fume hood above the phase-transition temperature. The dry lipid film was fully hydrated by vortexing to generate large liposomes. To prepare small liposomes, the hydrated lipids were sonicated in a bath for 1 h and extruded 35 times through a 0.1-μm polycarbonate membrane (Sigma-Aldrich, 610005) using an Avanti Mini-Extruder (Sigma-Aldrich, 610000). The quality and size of liposomes were assessed by dynamic light scattering using a DynaPro Titan dynamic light scattering instrument (Wyatt Technology), after which EM was conducted.
Liposome coflotation assay
Small liposomes were prepared using sonication and extrusion with a composition of 100 mol% DOPC or 30 mol% DPPS and 70 mol% DOPC. Small liposomes were incubated with 7 μM mouse TRIM72 WT and mutant proteins for 30 min at room temperature. The liposome–protein mixture was transferred to a centrifuge tube, after which sucrose solutions were overlaid on the mixture in the following order: 300 μl of 68% (wt/vol) sucrose, 2.5 ml of 17% (wt/vol) sucrose and 1 ml of 6.8% (wt/vol) sucrose in DPBS. The discontinuous sucrose gradient was centrifuged at 272,800g and 4 °C for 3 h. Each fraction from the top to the bottom of the gradient was resolved by SDS‒PAGE and analyzed by western blotting using a rabbit polyclonal anti-TRIM72 antibody (Abcam, ab154238).
Liposome co-sedimentation assay
Large liposomes with 80 mol% DOPC and 20 mol% cholesterol or 80 mol% PS and 20 mol% cholesterol were prepared by vortexing. Large liposomes were incubated with 50 μg of mouse TRIM72 WT or each mutant at room temperature for 30 min. The liposome–protein mixture was centrifuged at 21,130g and 4 °C for 10 min and then divided into supernatant and pellet fractions. Each fractionated sample was resolved by SDS‒PAGE, and signals were detected by Coomassie staining.
Flow cytometry
Fluorescence-labeled sfGFP–TRIM72 WT and FITC-labeled annexin V (FITC–annexin V; BioLegend, 640945) were prepared for flow cytometry analysis. Fluorescence-labeled and nascent liposomes were prepared by sonication in FACS buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM TCEP and 0.5% (wt/vol) BSA). Liposomes containing 99.9 mol% DOPC or 30 mol% DPPS and 69.9 mol% DOPC were labeled using 0.1 mol% 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (Sigma-Aldrich, 810150P). Rhodamine-labeled liposomes were analyzed using an Accuri C6 flow cytometer (BD Biosciences) with a 488-nm excitation laser and 533/30-nm (FL1; wavelength/filter) and 585/40-nm (FL2) emission detectors. The detection threshold was set to a forward scatter height of 2,000 and an FL2 height of 100. Spectral overlaps between sfGFP (or FITC) and rhodamine were compensated by sfGFP–TRIM72-WT-bound rhodamine-free 30 mol% PS liposomes and protein-free rhodamine-labeled 30 mol% PS liposomes. For analysis of Ca2+ effects, real-time flow cytometry was conducted with the following three sequential steps: (1) measurement of liposomes only, (2) measurement of liposomes to which protein (sfGFP–TRIM72 WT or FITC–annexin V) had been added and (3) measurement of liposomes and proteins to which CaCl2 (final concentration of 2.5 mM) had been added. Each step was conducted over a 5-min period, and the data were evaluated by using BD Accuri C6 Plus software (BD Biosciences).
Surface plasmon resonance
Surface plasmon resonance data were collected using a Biacore T200 instrument (Cytiva) with the Series S Sensor Chip L1 (Cytiva, 29104993). For immobilization, small liposomes were generated by sonication and extrusion. We calculated the sensorgrams upon subtraction of the signal for the immobilized small liposomes with 100 mol% DOPC (reference flow cell) from that for the immobilized small liposomes with 30 mol% DPPS and 70 mol% DOPC (second flow cell). To enhance immobilization efficiency, a 1:1 mixture of liposomes with 30 mol% PS and 2.5 M NaCl solution was injected into the flow cell. After immobilization, 0.2 mg ml−1 fatty acid-free BSA (Sigma-Aldrich, A7030) was used to block both liposome samples to prevent nonspecific binding. After normalization, serially diluted proteins were injected. Regeneration of the sensor chip was carried out by adding 50 mM NaOH solution between measurements. The binding kinetics were calculated using BIAevaluation (Cytiva) software.
Membrane-fractionation assay
C2C12 myoblasts were cultured in DMEM medium supplemented with 10% (vol/vol) FBS (Cytiva, SH30071.03IH25-40), penicillin (100 IU ml−1) and streptomycin (100 μg ml–1). Plasmids encoding 2× Strep–human TRIM72 WT and mutants were transfected into cells with the Neon transfection system (Thermo Fisher Scientific, MPL5000). After 48 h, cells were collected and separated into membrane and cytosolic fractions using a Mem-PER Plus Membrane fractionation kit (Thermo Fisher Scientific, 89842). Each fractionated sample (10 μg) was resolved by SDS‒PAGE and analyzed by western blotting using the following antibodies: anti-Strep (IBA, 2-1509-001), anti-caveolin-1 (CST, 3238), anti-GAPDH (Santa Cruz, sc-47724) and anti-β-actin (Santa Cruz, sc-47778).
Cross-linking assay
TRIM72-WT proteins were incubated with small liposomes composed of 80 mol% DOPS and 20 mol% cholesterol at room temperature for 3 h. After centrifugal filtration using Spin-X (Sigma-Aldrich, CLS8162), TRIM72 proteoliposomes were purified by SEC using Superose 6 Increase 10/300 GL resin in cross-linking buffer. The purified TRIM72 proteoliposomes were then incubated at 4 °C for 90 min with each of the following cross-linkers at 150 μM: bis(maleimido)ethane (BMOE; Thermo Fisher Scientific, 22323), 1,4-bismaleimidobutane (Thermo Fisher Scientific, 22331), 1,8-bismaleimido-diethyleneglycol (Thermo Fisher Scientific, 22336), 1,11-bismaleimido-triethyleneglycol (Thermo Fisher Scientific, 22337), bis(sulfosuccinimidyl)glutarate (ProteoChem, C1126) and bis(sulfosuccinimidyl)suberate (Thermo Fisher Scientific, 21580). The reactions were stopped by the addition of 25 mM Tris-HCl, pH 7.5, and 25 mM DTT and by boiling at 95 °C for 5 min with SDS sample buffer containing β-ME. The cross-linked products were analyzed by mass spectrometry (MS). For the time-dependent cross-linking reaction, mouse TRIM72 WT and mutants were preincubated with small liposomes containing 100 mol% DOPC or 30 mol% DPPS and 70 mol% DOPC in DPBS at room temperature for 30 min. The reactions were initiated by the addition of 20 μM BMOE to the protein–liposome mixtures at room temperature. After the cross-linking reaction was initiated by the addition of BMOE, it was stopped at each time point by the addition of 25 mM DTT and by boiling at 95 °C for 5 min with SDS sample buffer containing β-ME. Cross-linked proteins were resolved by SDS‒PAGE, and signals were detected by Coomassie staining.
Transmission electron microscopy examination of negative-stained samples
Human TRIM72 microvesicles were purified from HEK293T cells. Vesicle-bound and free-solution TRIM72 was copurified by AC using StrepTrap HP columns and fully separated by SEC (see details in the above Cloning, protein expression and purification section). To obtain mouse TRIM72 proteoliposomes, small liposomes with a composition of 30 mol% DPPS and 70 mol% DOPC were generated by sonication and extraction using 100-nm filters. The mixtures of small liposomes and purified mouse TRIM72-WT proteins were agitated gently at room temperature for 3 h to reconstitute TRIM72 proteoliposomes. The mouse TRIM72 proteoliposomes and free mouse TRIM72 proteins were further separated by SEC. The use of detergents was avoided to maintain the structure of microvesicles or liposomes. For negative EM, each peak fraction containing TRIM72 microvesicles or TRIM72 proteoliposomes was loaded onto glow-discharged carbon-coated grids (Electron Microscopy Sciences, CF200-Cu) and then rinsed and stained with 1% (wt/vol) uranyl acetate. Negative EM images were recorded on a charge-coupled device camera (2k by 2k, Gatan) using a Tecnai F20 microscope (FEI) operated at 200 kV.
Cryogenic electron tomography
For cryo-ET experiments, mouse TRIM72 proteoliposomes were reconstituted by incubation with mouse TRIM72 WT and small liposomes containing 80 mol% DOPS and 20 mol% cholesterol. The reconstituted TRIM72 proteoliposomes were further purified by SEC using cryo-ET buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl and 1 mM TCEP). The proteoliposomes were loaded onto glow-discharged holey grids (Quantifoil R1.2/1.3 300 Mesh, Copper; EMS, Q350CR1.3) and then vitrified using the Vitrobot Mark IV system (Thermo Fisher Scientific) at 4 °C and 90% relative humidity. Tilt series images were collected at the Korea Basic Science Institute using a Titan Krios G2 (Thermo Fisher Scientific) transmission electron microscope operated at 300 kV using a Falcon 3EC direct electron detector (Thermo Fisher Scientific) with Tomography 4.0 (Thermo Fisher Scientific) software, a tilt range from −60° to 60° and an angular increment of 2°. Images were recorded with defocus between −4.0 and −6.0 μm at a nominal magnification of ×47,000, which corresponds to a pixel size of 1.4 Å. The total dose per tomogram was approximately 200 e−/Å2. Tilt series alignment, contrast transfer function (CTF) correction and tomogram reconstruction were performed using the IMOD package90. Tomograms were reconstructed using weighted back projection (WBP) with a simultaneous iterative reconstruction technique (SIRT)-like filter and binned four times to a pixel size of 5.6 Å.
For subtomogram averaging of the mouse TRIM72-WT and TRIM72M138R-mutant proteoliposomes, we prepared large liposomes containing 80 mol% DOPS and 20 mol% cholesterol by vortexing. The large liposomes were incubated with mouse TRIM72 WT at 4 °C for 90 min. The mixtures of large liposomes and TRIM72 WT were fractionated by sucrose gradient centrifugation at 312,200g and 4 °C for 1 h. The cofloated fractions containing the TRIM72-WT proteoliposomes were collected and concentrated up to 0.1 mg ml−1. Due to spectral and colorimetric interference by phospholipid molecules, the protein concentration of the TRIM72 proteoliposomes was calculated from the comparison with the band intensities of quantified TRIM72 proteins. TRIM72M138R proteoliposomes were prepared by the liposome co-sedimentation protocol. Large liposomes were prepared by sonication in a bath for 1 h. The incubated mixtures with sonicated liposomes and TRIM72M138R proteins were centrifuged at 21,130g and 4 °C for 10 min. After the supernatant was removed, the pellet was washed three times and resuspended in cryo-ET buffer. TRIM72-WT and TRIM72M138R proteoliposomes were loaded onto glow-discharged holey grids (Quantifoil R1.2/1.3 300 Mesh, Copper; EMS, Q350CR1.3) and then vitrified using the Vitrobot Mark IV system at 4 °C and 90% relative humidity. To enhance the alignment of the tilt series images, AURION Gold Tracer (Aurion, 210.133) was used as a fiducial marker. Gold–BSA nanoparticles with a size of 10 nm were concentrated by collecting a pellet portion of the solution after centrifugation and then mixed with samples at a 1:5 (vol/vol) ratio just before the vitrification procedures. Tilt series images were collected using a Titan Krios G2 microscope operated at 300 kV equipped with a Falcon 3EC direct electron detector with an angular range from −54° to 54° and an angular increment of 3°. Images were recorded using defocus between −4.0 and −6.0 μm at a nominal magnification of ×29,000, which corresponded to a pixel size of 2.3 Å. The total dose per tomogram was approximately 52 e−/Å2. The tilt series were aligned using fiducial markers. Tomograms were reconstructed using SIRT and WBP, with 2× pixel binning and a pixel size of 4.6 Å. SIRT reconstruction was used for annotating subtomogram positions, whereas WBP reconstruction was used for subtomogram averaging.
Subtomogram averaging and model fitting
The particle coordinates were extracted from reconstructed tomograms of TRIM72-WT and TRIM72M138R proteoliposomes using the Dynamo package91. While assigning particle coordinates, the vesicle or ellipsoidal vesicle model was applied, depending on the shapes of the proteoliposomes. In total, we extracted 18,715 and 6,379 particle coordinates from the reconstructed tomograms of TRIM72-WT and TRIM72M138R proteoliposomes, respectively. The micrographs in the tilt series were subjected to beam-induced motion correction using MotionCor2 (ref. 92) followed by CTF estimation using CTFFIND4 (ref. 93). Subtomogram coordinates, tomogram alignment information and the estimated CTF were imported for subtomogram averaging using RELION-4.0 (ref. 94). An initial model was obtained from four binned pseudo-subtomograms with a box size of 96 pixels and a cropped box size of 48 pixels. The three-dimensional particles were aligned by carrying out a series of sequential steps of classification with alignment, refinement and further classification without alignment. Iterative refinements were performed while the micrographs were progressively unbinned to the original pixel size. After removing duplicates within 30 Å, CTF refinement and frame alignment were performed on the refined model with a box size of 512 pixels and a cropped box size of 192 pixels. The final resolutions of the reconstructions of TRIM72-WT and TRIM72M138R proteoliposomes were estimated to be 25 and 26 Å, respectively, based on an FSC of 0.143. The subtomogram averaging process is illustrated in Extended Data Fig. 8a. The statistics of cryo-ET are summarized in Table 2. The relative intensity of the projection of subtomogram averaging reconstruction was analyzed with Fiji95. The initial model of higher-order TRIM72 assembly on the lipid bilayer was derived from the crystal packing of mouse TRIM72 ΔRING. We docked the initial model into the cryo-ET maps by rigid-body fitting using UCSF ChimeraX87. The fitted model was further optimized by ‘phenix.dock_in_map’ (ref. 96). The model of the lipid bilayer was calculated by CHARMM-GUI97.
Cross-linking mass spectrometry
Each band corresponding to a cross-linked protein was subjected to in-gel digestion with trypsin for LC‒MS/MS analysis. LC‒MS/MS analysis was performed using a Dionex UltiMate 3000 nano HPLC system coupled online to a Q Exactive mass spectrometer (Thermo Fisher Scientific). Chromatographic separation was performed with a 150-mm × 75-μm Acclaim PepMap C18 reversed-phase analytical column (Dionex) using gradient elution (solvent A, 0.1% formic acid in water; solvent B, 0.1% formic acid in acetonitrile; 0–10 min, 2% B; 40 min, 30% B; 44–49 min, 90% B; 50–60 min, 2% B). MS data were acquired with the top ten data-dependent MS/MS scan methods. Raw data were converted to MGF format using MSConvert and used to search a database containing the mouse TRIM72 proteins with pLink 2 (ref. 98). Peptide spectrum matches were filtered at a false discovery rate of 1%. MS/MS spectra assigned to cross-linked peptides were further confirmed by manual validation. The distances between cross-linked residues in our structural models were calculated with Xwalk99.
Mass spectrometry
The fractions containing TRIM72 microvesicles copurified from mammalian cells were solubilized in lysis buffer containing 50 mM Tris-HCl, pH 8.0 and 4% (wt/vol) SDS with protease inhibitor cocktail. SDS lysates were digested using filter-aided sample preparation (FASP) as described previously100. LC‒MS/MS analysis was performed as described above in Cross-linking mass spectrometry. MS-derived data were used to search human entries in the UniProt database through the Proteome Discoverer platform (version 2.2.0.388, Thermo Fisher Scientific). The search parameters for the SEQUEST algorithm were as follows: carbamidomethylation of cysteine as a fixed modification, oxidation of methionine as a variable modification, a precursor mass tolerance of 10 ppm and a fragment mass tolerance of 0.02 Da. Peptide spectrum matches were validated using a percolator based on q values at a false discovery rate of 1%. Gene ontology analysis was performed using DAVID software to evaluate biological functions101. A P value < 0.05 was considered to indicate a statistically significant result.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 7XV2, 7XYY, 7XYZ, 7XZ0, 7XZ1 and 7XZ2 and in the Small Angle Scattering Biological Data Bank under accession codes SASDK86 and SASDK96. The subtomogram averages have been deposited in the Electron Microscopy Data Bank under accession codes EMD-31150, EMD-31151, EMD-33569 and EMD-33582. CLMS and gene ontology analysis have been deposited in the Proteomics Identifications Database under the accession codes PXD024946 and PXD024978. All other data and materials are available upon request from the corresponding author. Source data are provided with this paper.
References
Dias, C. & Nylandsted, J. Plasma membrane integrity in health and disease: significance and therapeutic potential. Cell Discov. 7, 4 (2021).
McNeil, P. L. & Kirchhausen, T. An emergency response team for membrane repair. Nat. Rev. Mol. Cell Biol. 6, 499–505 (2005).
Cooper, S. T. & McNeil, P. L. Membrane repair: mechanisms and pathophysiology. Physiol. Rev. 95, 1205–1240 (2015).
Liu, J. et al. Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat. Genet. 20, 31–36 (1998).
Sun, M. et al. Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum. Mol. Genet. 9, 2471–2478 (2000).
Huynh, C., Roth, D., Ward, D. M., Kaplan, J. & Andrews, N. W. Defective lysosomal exocytosis and plasma membrane repair in Chediak–Higashi/beige cells. Proc. Natl Acad. Sci. USA 101, 16795–16800 (2004).
Heilbrunn, L. The Dynamics of Living Protoplasm (Academic Press, 1956).
Andrews, N. W. & Corrotte, M. Plasma membrane repair. Curr. Biol. 28, R392–R397 (2018).
Zhen, Y., Radulovic, M., Vietri, M. & Stenmark, H. Sealing holes in cellular membranes. EMBO J. 40, e106922 (2021).
Lin, Y.-C., Chipot, C. & Scheuring, S. Annexin-V stabilizes membrane defects by inducing lipid phase transition. Nat. Commun. 11, 230 (2020).
Sønder, S. L. et al. Restructuring of the plasma membrane upon damage by LC3-associated macropinocytosis. Sci. Adv. 7, eabg1969 (2021).
Roman, W. et al. Muscle repair after physiological damage relies on nuclear migration for cellular reconstruction. Science 374, 355–359 (2021).
Gupta, S. et al. Lysosomal retargeting of Myoferlin mitigates membrane stress to enable pancreatic cancer growth. Nat. Cell Biol. 23, 232–242 (2021).
Ritter, A. T. et al. ESCRT-mediated membrane repair protects tumor-derived cells against T cell attack. Science 376, 377–382 (2022).
Huang, Y., Zhang, X., Wang, H.-W. & Yu, L. Assembly of Tetraspanin-enriched macrodomains contains membrane damage to facilitate repair. Nat. Cell Biol. 24, 825–832 (2022).
Cheng, X., Zhang, X., Yu, L. & Xu, H. Calcium signaling in membrane repair. Semin. Cell Dev. Biol. 45, 24–31 (2015).
Terasaki, M., Miyake, K. & McNeil, P. L. Large plasma membrane disruptions are rapidly resealed by Ca2+-dependent vesicle–vesicle fusion events. J. Cell Biol. 139, 63–74 (1997).
Cai, C. et al. MG53 nucleates assembly of cell membrane repair machinery. Nat. Cell Biol. 11, 56–64 (2009).
Lee, C. et al. TRIM72 negatively regulates myogenesis via targeting insulin receptor substrate-1. Cell Death Differ. 17, 1254–1265 (2010).
Wang, X. et al. Cardioprotection of ischemia/reperfusion injury by cholesterol-dependent MG53-mediated membrane repair. Circ. Res. 107, 76–83 (2010).
Duann, P. et al. MG53-mediated cell membrane repair protects against acute kidney injury. Sci. Transl. Med. 7, 279ra36 (2015).
Jia, Y. et al. Treatment of acute lung injury by targeting MG53-mediated cell membrane repair. Nat. Commun. 5, 4387 (2014).
Weisleder, N. et al. Recombinant MG53 protein modulates therapeutic cell membrane repair in treatment of muscular dystrophy. Sci. Transl. Med. 4, 139ra85 (2012).
Sermersheim, M. et al. MG53 suppresses interferon-β and inflammation via regulation of ryanodine receptor-mediated intracellular calcium signaling. Nat. Commun. 11, 3624 (2020).
Song, R. et al. Central role of E3 ubiquitin ligase MG53 in insulin resistance and metabolic disorders. Nature 494, 375–379 (2013).
Yi, J.-S. et al. MG53-induced IRS-1 ubiquitination negatively regulates skeletal myogenesis and insulin signalling. Nat. Commun. 4, 2354 (2013).
Reymond, A. et al. The tripartite motif family identifies cell compartments. EMBO J. 20, 2140–2151 (2001).
Meroni, G. & Diez‐Roux, G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays 27, 1147–1157 (2005).
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Buetow, L. & Huang, D. T. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 17, 626–642 (2016).
Deshaies, R. J. & Joazeiro, C. A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 (2009).
Meroni, G. in Proteostasis and Disease (eds Barrio, R. et al.) 311–325 (Springer, 2020).
Hatakeyama, S. TRIM proteins and cancer. Nat. Rev. Cancer 11, 792–804 (2011).
van Gent, M., Sparrer, K. M. & Gack, M. U. TRIM proteins and their roles in antiviral host defenses. Annu. Rev. Virol. 5, 385–405 (2018).
Massiah, M. A., Simmons, B. N., Short, K. M. & Cox, T. C. Solution structure of the RBCC/TRIM B-box1 domain of human MID1: B-box with a RING. J. Mol. Biol. 358, 532–545 (2006).
James, L. C., Keeble, A. H., Khan, Z., Rhodes, D. A. & Trowsdale, J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc. Natl Acad. Sci. USA 104, 6200–6205 (2007).
Park, E. Y. et al. Crystal structure of PRY–SPRY domain of human TRIM72. Proteins 78, 790–795 (2010).
Tsai, W.-W. et al. TRIM24 links a non-canonical histone signature to breast cancer. Nature 468, 927–932 (2010).
Sanchez, J. G. et al. The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer. Proc. Natl Acad. Sci. USA 111, 2494–2499 (2014).
Goldstone, D. C. et al. Structural studies of postentry restriction factors reveal antiparallel dimers that enable avid binding to the HIV-1 capsid lattice. Proc. Natl Acad. Sci. USA 111, 9609–9614 (2014).
Li, Y. et al. Structural insights into the TRIM family of ubiquitin E3 ligases. Cell Res. 24, 762–765 (2014).
Weinert, C., Morger, D., Djekic, A., Grütter, M. G. & Mittl, P. R. Crystal structure of TRIM20 C-terminal coiled-coil/B30. 2 fragment: implications for the recognition of higher order oligomers. Sci. Rep. 5, 10819 (2015).
Koliopoulos, M. G., Esposito, D., Christodoulou, E., Taylor, I. A. & Rittinger, K. Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity. EMBO J. 35, 1204–1218 (2016).
Wang, P. et al. RING tetramerization is required for nuclear body biogenesis and PML sumoylation. Nat. Commun. 9, 1277 (2018).
Koliopoulos, M. G. et al. Molecular mechanism of influenza A NS1-mediated TRIM25 recognition and inhibition. Nat. Commun. 9, 1820 (2018).
Dickson, C. et al. Intracellular antibody signalling is regulated by phosphorylation of the Fc receptor TRIM21. eLife 7, e32660 (2018).
Stoll, G. A. et al. Structure of KAP1 tripartite motif identifies molecular interfaces required for retroelement silencing. Proc. Natl Acad. Sci. USA 116, 15042–15051 (2019).
Lim, M. et al. A ubiquitin-binding domain that binds a structural fold distinct from that of ubiquitin. Structure 27, 1316–1325 (2019).
Kiss, L. et al. A tri-ionic anchor mechanism drives Ube2N-specific recruitment and K63-chain ubiquitination in TRIM ligases. Nat. Commun. 10, 4502 (2019).
Kato, K. et al. Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases. Mol. Cell 81, 599–613 (2021).
Ru, Y. et al. C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7. Proc. Natl Acad. Sci. USA 119, e2203218119 (2022).
Woo, J.-S., Suh, H.-Y., Park, S.-Y. & Oh, B.-H. Structural basis for protein recognition by B30.2/SPRY domains. Mol. Cell 24, 967–976 (2006).
Ikeda, K. & Inoue, S. in TRIM/RBCC Proteins 27–37 (Springer, 2012).
Rand, M. L. et al. Diannexin, an annexin A5 homodimer, binds phosphatidylserine with high affinity and is a potent inhibitor of platelet-mediated events during thrombus formation. J. Thromb. Haemost. 10, 1109–1119 (2012).
Lips, C. et al. Who with whom: functional coordination of E2 enzymes by RING E3 ligases during poly-ubiquitylation. EMBO J. 39, e104863 (2020).
Pruneda, J. N. et al. Structure of an E3:E2~Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol. Cell 47, 933–942 (2012).
Panjkovich, A. & Svergun, D. I. Deciphering conformational transitions of proteins by small angle X-ray scattering and normal mode analysis. Phys. Chem. Chem. Phys. 18, 5707–5719 (2016).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Swairjo, M. A., Concha, N. O., Kaetzel, M. A., Dedman, J. R. & Seaton, B. A. Ca2+-bridging mechanism and phospholipid head group recognition in the membrane-binding protein annexin V. Nat. Struct. Biol. 2, 968–974 (1995).
Bandi, S. R. et al. E3 ligase-defective Cbl mutants lead to a generalized mastocytosis and myeloproliferative disease. Blood 114, 4197–4208 (2009).
Kales, S. C., Ryan, P. E., Nau, M. M. & Lipkowitz, S. Cbl and human myeloid neoplasms: the Cbl oncogene comes of age. Cancer Res. 70, 4789–4794 (2010).
Branigan, E., Carlos Penedo, J. & Hay, R. T. Ubiquitin transfer by a RING E3 ligase occurs from a closed E2~ubiquitin conformation. Nat. Commun. 11, 2846 (2020).
Yudina, Z. et al. RING dimerization links higher-order assembly of TRIM5α to synthesis of K63-linked polyubiquitin. Cell Rep. 12, 788–797 (2015).
Dawidziak, D. M., Sanchez, J. G., Wagner, J. M., Ganser‐Pornillos, B. K. & Pornillos, O. Structure and catalytic activation of the TRIM23 RING E3 ubiquitin ligase. Proteins 85, 1957–1961 (2017).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Keown, J. R., Yang, J., Black, M. M. & Goldstone, D. C. The RING domain of TRIM69 promotes higher-order assembly. Acta Crystallogr. D 76, 954–961 (2020).
Wagner, J. M. et al. Mechanism of B-box 2 domain-mediated higher-order assembly of the retroviral restriction factor TRIM5α. eLife 5, e16309 (2016).
Biris, N. et al. Structure of the rhesus monkey TRIM5α PRYSPRY domain, the HIV capsid recognition module. Proc. Natl Acad. Sci. USA 109, 13278–13283 (2012).
Niu, Y. et al. Cryo-EM structure of human MG53 homodimer. Biochem. J. 479, 1909–1916 (2022).
Ma, Y., Ding, L., Li, Z. & Zhou, C. Structural basis for TRIM72 oligomerization during membrane damage repair. Nat. Commun. 14, 1555 (2023).
Pedelacq, J.-D., Cabantous, S., Tran, T., Terwilliger, T. C. & Waldo, G. S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006).
Berndsen, C. E. & Wolberger, C. A spectrophotometric assay for conjugation of ubiquitin and ubiquitin-like proteins. Anal. Biochem. 418, 102–110 (2011).
Jin, J., Li, X., Gygi, S. P. & Harper, J. W. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature 447, 1135–1138 (2007).
Sung, K. H. & Song, H. K. Direct recognition of the C-terminal polylysine residues of nonstop protein by Ltn1, an E3 ubiquitin ligase. Biochem. Biophys. Res. Commun. 453, 642–647 (2014).
Otwinowski, Z. & Minor, W. in Methods in Enzymology, Vol. 276 (eds Carter, C. W. & Sweet, R. M.) 307–326 (Academic Press, 1997).
Zwart, P. H. et al. Automated structure solution with the PHENIX suite. Methods Mol. Biol. 426, 419–435 (2008).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Brunger, A. T. et al. Application of DEN refinement and automated model building to a difficult case of molecular-replacement phasing: the structure of a putative succinyl-diaminopimelate desuccinylase from Corynebacterium glutamicum. Acta Crystallogr. D Biol. Crystallogr. 68, 391–403 (2012).
Freemont, P. S. RING for destruction? Curr. Biol. 10, R84–R87 (2000).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D 74, 531–544 (2018).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Pei, J., Kim, B.-H. & Grishin, N. V. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300 (2008).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Hall, T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 (1999).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Svergun, D. I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 (1999).
Dowler, S., Kular, G. & Alessi, D. R. Protein lipid overlay assay. Sci. STKE 2002, pl6 (2002).
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Castano-Diez, D., Kudryashev, M., Arheit, M. & Stahlberg, H. Dynamo: a flexible, user-friendly development tool for subtomogram averaging of cryo-EM data in high-performance computing environments. J. Struct. Biol. 178, 139–151 (2012).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Zivanov, J. et al. A Bayesian approach to single-particle electron cryo-tomography in RELION-4.0. eLife 11, e8372 (2022).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D 75, 861–877 (2019).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Chen, Z.-L. et al. A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nat. Commun. 10, 3404 (2019).
Kahraman, A., Malmstrom, L. & Aebersold, R. Xwalk: computing and visualizing distances in cross-linking experiments. Bioinformatics 27, 2163–2164 (2011).
Ludwig, K. R., Schroll, M. M. & Hummon, A. B. Comparison of in-solution, FASP, and S-Trap based digestion methods for bottom–up proteomic studies. J. Proteome Res. 17, 2480–2490 (2018).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Rambo, R. P. & Tainer, J. A. Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496, 477–481 (2013).
Acknowledgements
We thank the staff at beamlines 4C, 5C and 11C at the Pohang Accelerator Laboratory in Korea and beamlines BL-17A and BL-10C at the Photon Factory in Japan for help with X-ray data collection. This work was performed in part under the International Collaborative Research Program of the Institute for Protein Research, Osaka University (ICR-21-05). Diffraction data were collected at Osaka University beamline BL44XU at SPring-8 (Harima, Japan) (proposal nos. 2021A6673 and 2021B6673). E. Yamashita at Osaka University provided helpful discussion during crystallographic data analysis. We thank M. H. Nam at the Korea Basic Science Institute, Seoul Center, for her help with Biacore experiments and N. Thomä at the Friedrich Miescher Institute for Biomedical Research for his comments on the paper. This study was supported by National Research Foundation of Korea grants from the Korean government (grant nos. 2020R1A2C3008285, 2021M3A9I4030068 and 2022M3A9G8082638).
Author information
Authors and Affiliations
Contributions
S.H.P., J. Han and B.-C.J. performed biochemical experiments. S.H.P., J. Han, B.-C.J. and B.H.K. purified recombinant proteins. S.H.P. prepared crystals, solved structures and conducted SAXS experiments. S.H.P. and K.E.L. performed TEM studies. S.H.P., J. Han and J.H.S. performed cell biology experiments. S.H.P., S.H.J. and Z.-Y.P. performed mass spectrometric analysis. S.H.P., H.J. and J. Hyun performed cryo-ET experiments. S.H.P., K.E.L., Y.-G.K., J. Hyun and H.K.S. analyzed data. S.H.P. and H.K.S. designed experiments and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Sun Hur and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Katarzyna Ciazynska, in collaboration with the Nature Structural & Molecular Biology team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 SEC-MALS and SEC-SAXS analysis of TRIM72.
a, SDS‒PAGE analysis of TRIM72 WT and ΔRING. All proteins used in this study were purified to equivalent purity. b, SEC-MALS profiles of TRIM72 WT at various concentrations. The relative Rayleigh ratios (left axis) and molecular masses (right axis) are shown as dashed lines and open circles of the same color for each concentration, respectively (11.2 mg/ml, red; 5.6 mg/ml, blue; 2.8 mg/ml, green). c-d, SEC-SAXS profiles of TRIM72 WT (c) and ΔRING (d). The average intensity (left axis) is shown as a dashed line. The values (right axis) with volume of correction (Vc)102 and radius of gyration (Rg) are indicated by open circles in green and red, respectively. e–f, Scattering intensity, Guinier and Kratky plots, and paired distance distribution function P(r) of TRIM72 WT (e) and ΔRING (f). All SAXS analyses were performed using the average of the peak fractions of SEC-SAXS for each protein.
Extended Data Fig. 2 Comparison of the ligand binding sites among PRYSPRY domains.
a-f, Structural comparison of PRYSPRY domains. Mouse TRIM72 PRYSPRY in dimer (a), rhesus TRIM5α PRYSPRY (b), human TRIM7 PRYSPRY complexed with 2 C peptides of human enterovirus (c), human TRIM21 PRYSPRY complexed with human IgG Fc (d), human TRIM65 PRYSPRY complexed with human MDA5 helicase domain (e), and human RIPLET PRYSPRY complexed with the human RIG-I helicase domain (f). The PRYSPRY domains are shown in a ribbon diagram and electrostatic surface model with transparency. The binding molecules within 10 Å distances of PRYSPRY are shown in a ribbon diagram colored yellow. The PRYSPRY domain of mouse TRIM72 was derived from the crystal structure of TRIM72 ΔRING (2.75 Å). g, Structure-based sequence alignments of PRYSPRY domains corresponding to a-f. Mutated residues of mouse TRIM72 in this study are shown in red boxes. Potentially essential residues of TRIM72 in membrane binding are indicated as pink boxes. The proposed residues of rhesus TRIM5α PRYSPRY for recognizing HIV capsids as indicated by NMR and HIV-1 restriction assays are shown in orange boxes. Pro334 in rhesus TRIM5α (Arg332 in humans), which is a critical residue for HIV restriction, is highlighted with a yellow box. The important residues of human TRIM7 PRYSPRY for recognizing C2 peptides from human enterovirus are shown in magenta boxes. Four hotspot residues of human TRIM21 PRYSPRY in contact with human IgG Fc are shown in mint boxes. The critical residues in human TRIM65 PRYSPRY for IFNβ mRNA levels upon poly (I:C) stimulation are shown in cyan boxes. The important residues in human RIPLET PRYSPRY for IFNβ mRNA levels upon stimulation with dsRNA are shown in green boxes. All boxed residues are represented as stick models in a-f. The variable loop (VL) regions 1-6 are indicated as black-lined boxes. Note that the critical residues are crowded in VL1, VL3, VL4, and VL6. The mutated residues in the H3 helix for crystallization of mouse TRIM72 (K279H/A283H) are shown in blue.
Extended Data Fig. 3 Protein–lipid overlay analysis.
a-d, Protein–lipid overlay assay using PIP StripsTM (a), Inositol Snoopers® (b), Oxidized Phospholipid Snoopers® (c) and Sphingolipid Snoopers® (d) in the absence (left) or presence (middle) of Ca2+. The spotted lipid positions are indicated in the right panel. e, Lists of the examined lipids. TRIM72 WT was incubated and detected with a rabbit polyclonal anti-TRIM72 antibody. The detailed experimental procedures are described in the Methods.
Extended Data Fig. 4 SPR analysis of TRIM72 WT and variants.
a, Binding affinity KD values of TRIM72 WT and variants to PS-liposomes. n.d., not determined. b-h, Sensorgrams with different concentrations of TRIM72 WT (b), K2D (c), R3E (d), ΔH3 (e), RBCC (f), GST-H3-PRYSPRY (g), and MBP-H3-PRYSPRY (h) are shown. The first and second vertical dashed lines indicate the start and end of the injection, respectively. The PS-liposomes contained 30 mol% PS. The detailed experimental procedures are described in the Methods.
Extended Data Fig. 5 Crosslinking assays to identify the TRIM72 oligomer.
a, Overall scheme of the time-dependent crosslinking assay. b-e, Time-dependent crosslinking analysis of mouse TRIM72 WT (b) and variants (c-e). Note that each variant of K2D (c), R3E (d), and ΔH3 (e) forms almost no high-molecular-weight crosslinked oligomers. All PS-liposomes contained 30 mol% PS.
Extended Data Fig. 6 Higher-order TRIM72 assembly on both microvesicles and liposomes.
a, Time-dependent crosslinking analysis of TRIM72 WT purified from mammalian cells. b, SEC profiles of TRIM72 expressed in mammalian cells (blue line) and in vitro-reconstituted TRIM72 proteoliposomes (red line). The fractions containing TRIM72 bound to microvesicles or liposomes and TRIM72 in solution are indicated by arrows colored green and pink, respectively. c, Negative-stained TEM images of TRIM72 bound to microvesicles at low (left) and high magnifications (right). d, TEM images of the negatively stained TRIM72 proteoliposomes. (c, d) Higher-order TRIM72 assemblies are indicated by black arrowheads. The red and black scale bars indicate 100 and 20 nm, respectively. e, Cryo-electron tomograms (left) visualized by 3D volume rendering (right) showing the higher-order TRIM72 assembly on the inside convex (up) and outside concave (down) sides of small liposomes. The TRIM72 assembly and liposomal membranes are colored cyan and white, respectively. The black scale bars indicate 20 nm in e.
Extended Data Fig. 7 Subtomogram averaging reconstruction of the higher-order TRIM72 WT assembly.
a, Data processing of subtomogram averaging of the TRIM72 assembly. b, Angular distribution of the final refined map. c, Contour analysis of TRIM72 assembly. Note that the higher-order TRIM72 assembly is clearly distinguished from the phospholipid bilayer. The black scale bars indicate 100 nm (b, c). d, Fourier shell correlation (FSC) curve of the final refined subtomogram averaging map of the TRIM72 WT assembly. FSC curve calculated from two independently refined half-maps indicating the overall estimated resolution at 25 Å based on a 0.143 FSC criterion. The cryoET statistics of the TRIM72 WT assembly are summarized in Table 2.
Extended Data Fig. 8 CryoET and subtomogram averaging reconstruction of the higher-order TRIM72 M138R assembly.
a, Liposome cosedimentation analysis. b, Micrograph of the TRIM72 M138A proteoliposome. c, Tomogram slices of TRIM72 WT (left) and M138R proteoliposomes (right). The black boxes in the top panels are enlarged in the bottom panels. Note that the TRIM72 assemblies are shown in both TRIM72 WT and M138R but not in M138A. The black and white scale bars indicate 100 nm and 20 nm, respectively. (b,c). d, Subtomogram averaging reconstruction of TRIM72 M138R. The averages of Z-slices correspond to the middle (left) and front view (middle). The relative intensity according to the x-axis (left) was calculated from the averaged Z-slices of the front view. The number in italics (1-5) indicates each column-like density. The regular intervals between the peak density are 6 nm, in agreement with the intervals observed for the TRIM72 WT assembly. The black scale bar indicates 5 nm in d. e, Angular distribution of the final refined map. f, higher-order TRIM72 M138R assembly fitted into the subtomogram averaging map. The black scale bars indicate 100 nm (e,f). g, FSC curve of the final refined subtomogram averaging map of the TRIM72 M138R assembly. FSC curve calculated from two independently refined half-maps indicating overall resolution at 26 Å based on the 0.143 FSC criterion. The cryoET statistics are summarized in Table 2.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2 and Figs. 1–9.
Supplementary Data 1–3
Supplementary Data 1: Proteomic analysis of TRIM72-containing microvesicles. Supplementary Data 2: CLMS analysis. Supplementary Data 3: Analysis of linchpin residues in E3 Ub ligases.
Supplementary Data 4
Predicted structures of the RING domain of 60 TRIM proteins.
Supplementary Video 1
Slicing in z through a tomogram showing the convex surface of liposomes.
Supplementary Video 2
Slicing in z through a tomogram showing the concave surface of liposomes.
Supplementary Video 3
Slicing in z through a tomogram of TRIM72 WT on liposomes used for subtomogram averaging.
Supplementary Video 4
Slicing in z through a tomogram of the TRIM72M138R mutant on liposomes used for subtomogram averaging.
Supplementary Data 5
Unprocessed western blots for Supplementary Figs. 4, 8a–d and 9d.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Source data for analyzing the assembly pattern.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Source data for SEC–MALS analysis.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 4
Source data for surface plasmon resonance.
Source Data Extended Data Fig. 6
Source data for SEC–MALS analysis.
Source Data Extended Data Fig. 7
Source data for the FSC curve.
Source Data Extended Data Fig. 8
Source data for the FSC curve.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Park, S.H., Han, J., Jeong, BC. et al. Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane. Nat Struct Mol Biol 30, 1695–1706 (2023). https://doi.org/10.1038/s41594-023-01111-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-023-01111-7