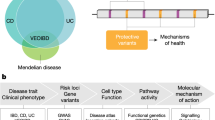
Abstract
Crohn's disease and ulcerative colitis are prototypical complex diseases characterized by chronic and heterogeneous manifestations, induced by interacting environmental, genomic, microbial and immunological factors. These interactions result in an overwhelming complexity that cannot be tackled by studying the totality of each pathological component (an '–ome') in isolation without consideration of the interaction among all relevant –omes that yield an overall 'network effect'. The outcome of this effect is the 'IBD interactome', defined as a disease network in which dysregulation of individual –omes causes intestinal inflammation mediated by dysfunctional molecular modules. To define the IBD interactome, new concepts and tools are needed to implement a systems approach; an unbiased data-driven integration strategy that reveals key players of the system, pinpoints the central drivers of inflammation and enables development of targeted therapies. Powerful bioinformatics tools able to query and integrate multiple –omes are available, enabling the integration of genomic, epigenomic, transcriptomic, proteomic, metabolomic and microbiome information to build a comprehensive molecular map of IBD. This approach will enable identification of IBD molecular subtypes, correlations with clinical phenotypes and elucidation of the central hubs of the IBD interactome that will aid discovery of compounds that can specifically target the hubs that control the disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Busse, R., Blu¨mel, M., Scheller-Kreinsen, D. & Zentner, A. Tackling Chronic Diseases in Europe — Strategies, Interventions and Challenges. (European Observatory on Health Systems and Polices & World Health Organization, 2010).
Iyengar, R. Complex diseases require complex therapies. EMBO Rep. 14, 1039–1042 (2013). This insightful commentary highlights the molecular complexity of chronic diseases and suggests that current therapeutic approaches may have reached their limits.
Sivakumaran, S. et al. Abundant pleiotropy in human complex diseases and traits. Am. J. Hum. Genet. 89, 607–618 (2011).
Ghiassian, S. D. et al. Endophenotype network models: common core of complex diseases. Sci. Rep. 6, 27414 (2016). This article presents an alternative view emphasizing that different diseases often have common underlying mechanisms and shared endophenotypes. The authors construct endophenotype network models and explore their relation to different diseases, identifying critical modules within the subnetworks of the human interactome.
de Souza, H. S. & Fiocchi, C. Immunopathogenesis of IBD: current state of the art. Nat. Rev. Gastroenterol. Hepatol. 13, 13–27 (2016). The authors present the current knowledge of the immunity and expand the concept of IBD immunopathogenesis to include various novel components that affect the intestinal inflammatory process.
Rogler, G. & Vavricka, S. Exposome in IBD: recent insights in environmental factors that influence the onset and course of IBD. Inflamm. Bowel Dis. 21, 400–408 (2015).
Saidel-Odes, L. & Odes, S. Hygiene hypothesis in inflammatory bowel disease. Ann. Gastroenterol. 27, 189–190 (2014).
Cleynen, I. & Vermeire, S. The genetic architecture of inflammatory bowel disease: past, present and future. Curr. Opin. Gastroenterol. 31, 456–463 (2015).
McGovern, D. P., Kugathasan, S. & Cho, J. H. Genetics of inflammatory bowel diseases. Gastroenterology 149, 1163–1176.e2 (2015).
Wang, M. H. et al. A novel approach to detect cumulative genetic effects and genetic interactions in Crohn's disease. Inflamm. Bowel Dis. 19, 1799–1808 (2013).
Stappenbeck, T. S. et al. Crohn disease: a current perspective on genetics, autophagy and immunity. Autophagy 7, 355–374 (2011).
Kostic, A. D., Xavier, R. J. & Gevers, D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146, 1489–1499 (2014).
Manichanh, C., Borruel, N., Casellas, F. & Guarner, F. The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 9, 599–608 (2012).
Choung, R. S. et al. Serologic microbial associated markers can predict Crohn's disease behaviour years before disease diagnosis. Aliment. Pharmacol. Ther. 43, 1300–1310 (2016).
Darfeuille-Michaud, A. et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 127, 412–421 (2004).
Machiels, K. et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63, 1275–1283 (2013).
Goodrich, J. K. et al. Human genetics shape the gut microbiome. Cell 159, 789–799 (2014).
Braunstein, J., Qiao, L., Autschbach, F., Schurmann, G. & Meuer, S. C. T cells of the human intestinal lamina propria are high producers of interleukin 10. Gut 53, 215–220 (1997).
Monteleone, G. et al. Blocking Smad7 restores TGF-β1 signaling in chronic inflammatory bowel disease. J. Clin. Invest. 108, 601–609 (2001).
Neurath, M. F. Cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 14, 329–342 (2014).
Reinisch, W. et al. A dose escalating, placebo controlled, double blind, single dose and multidose, safety and tolerability study of fontolizumab, a humanised anti-interferon gamma antibody, in patients with moderate to severe Crohn's disease. Gut 55, 1138–1144 (2006).
Hueber, W. et al. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn's disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut 61, 1693–1700 (2012).
Danese, S. et al. Tralokinumab for moderate-to-severe UC: a randomised, double-blind, placebo-controlled, phase IIa study. Gut 64, 243–249 (2015).
Spencer, D. M., Veldman, G. M., Banerjee, S., Willis, J. & Levine, A. D. Distinct inflammatory mechanisms mediate early versus late colitis in mice. Gastroenterology 122, 94–105 (2002).
Kugathasan, S. et al. Mucosal T-cell immunoregulation varies in early and late Crohn's disease. Gut 56, 1696–1705 (2007).
Wang, P., Han, W. & Ma, D. Electronic sorting of immune cell subpopulations based on highly plastic genes. J. Immunol. 197, 665–673 (2016).
Berg, J. Genes-environment interplay. Science 354, 15 (2016).
Fan, S., Hansen, M. E., Lo, Y. & Tishkoff, S. A. Going global by adapting local: A review of recent human adaptation. Science 354, 54–59 (2016).
Miska, E. A. & Ferguson-Smith, A. C. Transgenerational inheritance: Models and mechanisms of non-DNA sequence-based inheritance. Science 354, 59–63 (2016).
Yosef, N. & Regev, A. Writ large: Genomic dissection of the effect of cellular environment on immune response. Science 354, 64–68 (2016).
Franks, P. W. & McCarthy, M. I. Exposing the exposures responsible for type 2 diabetes and obesity. Science 354, 69–73 (2016).
Suez, J. et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature 514, 181–186 (2014).
Kreitinger, J. M., Beamer, C. A. & Shepherd, D. M. Environmental immunology: lessons learned from exposure to a select panel of immunotoxicants. J. Immunol. 196, 3217–3225 (2016).
Carr, E. J. et al. The cellular composition of the human immune system is shaped by age and cohabitation. Nat. Immunol. 17, 461–468 (2016). Using a systems-level approach, the authors establish a resource of cellular immune profiles of healthy individuals.
Ye, B. D. & McGovern, D. P. Genetic variation in IBD: progress, clues to pathogenesis and possible clinical utility. Expert Rev. Clin. Immunol. 12, 1091–1107 (2016).
Glocker, E. O. et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 361, 2033–2045 (2009).
Chakravarti, A. Genomics is not enough. Science 334, 15 (2011).
Abreu, M. T. et al. Mutations in NOD2 are associated with fibrostenosing disease in patients with Crohn's disease. Gastroenterology 123, 679–688 (2002).
Sahni, N. et al. Widespread macromolecular interaction perturbations in human genetic disorders. Cell 161, 647–660 (2015).
Chen, Y. et al. Variations in DNA elucidate molecular networks that cause disease. Nature 452, 429–435 (2008).
Benfey, P. N. & Mitchell-Olds, T. From genotype to phenotype: systems biology meets natural variation. Science 320, 495–497 (2008). The authors propose that systems biology approaches can be applied to decipher gene's function and related abnormalities, generating genotype-phenotype maps.
Manolio, T. A. Genomewide association studies and assessment of the risk of disease. N. Engl. J. Med. 363, 166–176 (2010).
Silvestri, G. A. et al. A bronchial genomic classifier for the diagnostic evaluation of lung cancer. N. Engl. J. Med. 373, 243–251 (2015).
Bailey, P. et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52 (2016).
Papaemmanuil, E. et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 374, 2209–2221 (2016).
Ellison, D. W. Multiple molecular data sets and the classification of adult diffuse gliomas. N. Engl. J. Med. 372, 2555–2557 (2015).
Edwards, S. L., Beesley, J., French, J. D. & Dunning, A. M. Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 93, 779–797 (2013).
[No authors listed.] Editorial: beyond the genome. Nature 518, 273 (2015).
Kasowski, M. et al. Extensive variation in chromatin states across humans. Science 342, 750–752 (2013).
Kundaje, A. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). In this study, investigators establish global maps of regulatory elements and define regulatory modules, revealing biologically relevant cell types for diverse human traits, and demonstrating the central role of epigenomic events for the development of disease.
Ventham, N. T., Kennedy, N. A., Nimmo, E. R. & Satsangi, J. Beyond gene discovery in inflammatory bowel disease: the emerging role of epigenetics. Gastroenterology 145, 293–308 (2013).
Nimmo, E. R. et al. Genome-wide methylation profiling in Crohn's disease identifies altered epigenetic regulation of key host defense mechanisms including the Th17 pathway. Inflamm. Bowel Dis. 18, 889–899 (2012).
Polytarchou, C. et al. MicroRNA214 is associated with progression of ulcerative colitis, and inhibition reduces development of colitis and colitis-associated cancer in mice. Gastroenterology 149, 981–992.e11 (2015).
Liu, S. et al. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe 19, 32–43 (2016).
Lee, T. I. & Young, R. A. Transcriptional regulation and its misregulation in disease. Cell 152, 1237–1251 (2013).
Planell, N. et al. Transcriptional analysis of the intestinal mucosa of patients with ulcerative colitis in remission reveals lasting epithelial cell alterations. Gut 62, 967–976 (2013).
Li, M. et al. Widespread RNA and DNA sequence differences in the human transcriptome. Science 333, 53–58 (2011).
Wu, L. et al. Variation and genetic control of protein abundance in humans. Nature 499, 79–82 (2013).
Starr, A. E. et al. Proteomic analysis of ascending colon biopsies from a paediatric inflammatory bowel disease inception cohort identifies protein biomarkers that differentiate Crohn's disease from UC. Gut http://dx.doi.org/10.1136/gutjnl-2015-310705 (2016).
Schicho, R. et al. Quantitative metabolomic profiling of serum, plasma, and urine by 1H NMR spectroscopy discriminates between patients with inflammatory bowel disease and healthy individuals. J. Proteome Res. 11, 3344–3357 (2012).
Patti, G. J., Yanes, O. & Siuzdak, G. Innovation: metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 13, 263–269 (2012).
Lim, E. S. et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat. Med. 21, 1228–1234 (2015).
Zhernakova, A. et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352, 565–569 (2016).
Franzosa, E. A. et al. Identifying personal microbiomes using metagenomic codes. Proc. Natl Acad. Sci. USA 112, E2930–E2938 (2015).
Chu, H. et al. Gene–microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 352, 1116–1120 (2016).
Imhann, F. et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut http://dx.doi.org/10.1136/gutjnl-2016-312135 (2016).
Van Noorden, R. Global scientific output doubles every nine years. Nature News Blog http://blogs.nature.com/news/2014/05/global-scientific-output-doubles-every-nine-years.html(2014).
Goldberger, A. L. Non-linear dynamics for clinicians: chaos theory, fractals, and complexity at the bedside. Lancet 347, 1312–1314 (1996).
West, B. J. A mathematics for medicine: the Network Effect. Front. Physiol. 5, 456 (2014).
Sanchez, C. et al. Grasping at molecular interactions and genetic networks in Drosophila melanogaster using FlyNets, an Internet database. Nucleic Acids Res. 27, 89–94 (1999).
Barabasi, A. L. & Albert, R. Emergence of scaling in random networks. Science 286, 509–512 (1999).
Ideker, T., Galitski, T. & Hood, L. A new approach to decoding life: systems biology. Annu. Rev. Genomics Hum. Genet. 2, 343–372 (2001). A very comprehensive review of the concept and application of systesm biology in biomedicine.
Polytarchou, C., Koukos, G. & Iliopoulos, D. Systems biology in inflammatory bowel diseases: ready for prime time. Curr. Opin. Gastroenterol. 30, 339–346 (2014). A systems biology approach, including novel computational methodologies capable of integrating high-throughput molecular data, is proposed for the study of pathogenic mechanisms in IBD.
Kabakchiev, B. & Silverberg, M. S. Expression quantitative trait loci analysis identifies associations between genotype and gene expression in human intestine. Gastroenterology 144, 1488–1496.e3 (2013).
Adams, A. T. et al. Two-stage genome-wide methylation profiling in childhood-onset Crohn's Disease implicates epigenetic alterations at the VMP1/MIR21 and HLA loci. Inflamm. Bowel Dis. 20, 1784–1793 (2014).
Saito, S. et al. DNA methylation of colon mucosa in ulcerative colitis patients: correlation with inflammatory status. Inflamm. Bowel Dis. 17, 1955–1965 (2011).
Foran, E. et al. Upregulation of DNA methyltransferase-mediated gene silencing, anchorage-independent growth, and migration of colon cancer cells by interleukin-6. Mol. Cancer Res. 8, 471–481 (2010).
Jones, B. & Chen, J. Inhibition of IFN-gamma transcription by site-specific methylation during T helper cell development. EMBO J. 25, 2443–2452 (2006).
Gu, H. et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat. Protoc. 6, 468–481 (2011).
Chen, T. & Dent, S. Y. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat. Rev. Genet. 15, 93–106 (2014).
Zhang, Q. et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature 525, 389–393 (2015).
Meddens, C. A. et al. Systematic analysis of chromatin interactions at disease associated loci links novel candidate genes to inflammatory bowel disease. Genome Biol. 17, 247 (2016).
Ellinghaus, D. et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat. Genet. 48, 510–518 (2016).
Koukos, G. et al. MicroRNA-124 regulates STAT3 expression and is down-regulated in colon tissues of pediatric patients with ulcerative colitis. Gastroenterology 145, 842–852.e2 (2013).
Koukos, G. et al. A microRNA signature in pediatric ulcerative colitis: deregulation of the miR-4284/CXCL5 pathway in the intestinal epithelium. Inflamm. Bowel Dis. 21, 996–1005 (2015).
Wu, F. et al. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology 135, 1624–1635.e24 (2008).
Davidovich, C. & Cech, T. R. The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA 21, 2007–2022 (2015).
Lasda, E. & Parker, R. Circular RNAs: diversity of form and function. RNA 20, 1829–1842 (2014).
M'Koma, A. E. et al. Proteomic profiling of mucosal and submucosal colonic tissues yields protein signatures that differentiate the inflammatory colitides. Inflamm. Bowel Dis. 17, 875–883 (2011).
Meuwis, M. A. et al. Proteomics for prediction and characterization of response to infliximab in Crohn's disease: a pilot study. Clin. Biochem. 41, 960–967 (2008).
Humphrey, S. J., Azimifar, S. B. & Mann, M. High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat. Biotechnol. 33, 990–995 (2015).
Le Gall, G. et al. Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J. Proteome Res. 10, 4208–4218 (2011).
Stephens, N. S. et al. Urinary NMR metabolomic profiles discriminate inflammatory bowel disease from healthy. J. Crohns Colitis 7, e42–e48 (2013).
Lupp, C. et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 119–129 (2007).
Ohkusa, T. et al. Fusobacterium varium localized in the colonic mucosa of patients with ulcerative colitis stimulates species-specific antibody. J. Gastroenterol. Hepatol. 17, 849–853 (2002).
Ogura, Y. et al. A frameshift mutation in Nod2 associated with susceptibility to Crohn's disease. Nature 411, 603–606 (2001).
Gedela, S. Integration, warehousing, and analysis strategies of Omics data. Methods Mol. Biol. 719, 399–414 (2011).
Shen, R., Olshen, A. B. & Ladanyi, M. Integrative clustering of multiple genomic data types using a joint latent variable model with application to breast and lung cancer subtype analysis. Bioinformatics 25, 2906–2912 (2009).
Shen, R. et al. Integrative subtype discovery in glioblastoma using iCluster. PLoS ONE 7, e35236 (2012).
Mo, Q. et al. Pattern discovery and cancer gene identification in integrated cancer genomic data. Proc. Natl Acad. Sci. USA 110, 4245–4250 (2013).
Vaske, C. J. et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics 26, i237–245 (2010).
Paull, E. O. et al. Discovering causal pathways linking genomic events to transcriptional states using Tied Diffusion Through Interacting Events (TieDIE). Bioinformatics 29, 2757–2764 (2013).
Wang, W. et al. iBAG: integrative Bayesian analysis of high-dimensional multiplatform genomics data. Bioinformatics 29, 149–159 (2013).
Calvano, S. E. et al. A network-based analysis of systemic inflammation in humans. Nature 437, 1032–1037 (2005).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Saito, R. et al. A travel guide to Cytoscape plugins. Nat. Methods 9, 1069–1076 (2012).
Maere, S., Heymans, K. & Kuiper, M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 (2005).
Vlasblom, J. et al. GenePro: a Cytoscape plug-in for advanced visualization and analysis of interaction networks. Bioinformatics 22, 2178–2179 (2006).
Xia, T. & Dickerson, J. A. OmicsViz: Cytoscape plug-in for visualizing omics data across species. Bioinformatics 24, 2557–2558 (2008).
Gao, J. et al. Metscape: a Cytoscape plug-in for visualizing and interpreting metabolomic data in the context of human metabolic networks. Bioinformatics 26, 971–973 (2010).
Moni, M. A., Xu, H. & Lio, P. CytoCom: a Cytoscape app to visualize, query and analyse disease comorbidity networks. Bioinformatics 31, 969–971 (2015).
Hu, Z., Mellor, J., Wu, J. & DeLisi, C. VisANT: an online visualization and analysis tool for biological interaction data. BMC Bioinformatics 5, 17 (2004).
Nikitin, A., Egorov, S., Daraselia, N. & Mazo, I. Pathway studio — the analysis and navigation of molecular networks. Bioinformatics 19, 2155–2157 (2003).
Iragne, F., Nikolski, M., Mathieu, B., Auber, D. & Sherman, D. ProViz: protein interaction visualization and exploration. Bioinformatics 21, 272–274 (2005).
Agoston, V., Cemazar, M., Kajan, L. & Pongor, S. Graph-representation of oxidative folding pathways. BMC Bioinformatics 6, 19 (2005).
Pujol, A., Mosca, R., Farres, J. & Aloy, P. Unveiling the role of network and systems biology in drug discovery. Trends Pharmacol. Sci. 31, 115–123 (2010).
Lamb, J. et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006).
Kutalik, Z., Beckmann, J. S. & Bergmann, S. A modular approach for integrative analysis of large-scale gene-expression and drug-response data. Nat. Biotechnol. 26, 531–539 (2008).
Ma, H. & Zhao, H. iFad: an integrative factor analysis model for drug-pathway association inference. Bioinformatics 28, 1911–1918 (2012).
Shankavaram, U. T. et al. CellMiner: a relational database and query tool for the NCI-60 cancer cell lines. BMC Genomics 10, 277 (2009).
Woo, J. H. et al. Elucidating compound mechanism of action by network perturbation analysis. Cell 162, 441–451 (2015).
Hu, Z. et al. VisANT 4.0: integrative network platform to connect genes, drugs, diseases and therapies. Nucleic Acids Res. 41, W225–W231 (2013).
Hirsch, H. A., Iliopoulos, D., Tsichlis, P. N. & Struhl, K. Metformin selectively targets cancer stem cells, and acts together with chemotherapy to block tumor growth and prolong remission. Cancer Res. 69, 7507–7511 (2009).
Iliopoulos, D., Hirsch, H. A. & Struhl, K. Metformin decreases the dose of chemotherapy for prolonging tumor remission in mouse xenografts involving multiple cancer cell types. Cancer Res. 71, 3196–3201 (2011).
Niraula, S. et al. Metformin in early breast cancer: a prospective window of opportunity neoadjuvant study. Breast Cancer Res. Treat. 135, 821–830 (2012).
Dudley, J. T. et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci. Transl Med. 3, 96ra76 (2011).
Loscalzo, J., Barabasi, A. L. & Silverman, E. K. Network Medicine: Complex Systems in Human Disease and Therapeutics. (Harvard Univ. Press, 2017). This book introduces and explains the concept of network medicine as the correct approach to tackle the molecular underpinnings of most complex diseases. Various chapters address molecular, translational and clinical aspects of network medicine in a fashion accessible to non-experts.
Li-Pook-Than, J. & Snyder, M. iPOP goes the world: integrated personalized Omics profiling and the road toward improved health care. Chem. Biol. 20, 660–666 (2013).
Banchereau, R. et al. Personalized immunomonitoring uncovers molecular networks that stratify lupus patients. Cell 165, 551–565 (2016). In this study, the molecular heterogeneity of systemic lupus erythematosus, a complex autoimmune disease, is revealed through a systems biology approach, offering an explanation for the failure of current clinical trials.
Jameson, J. L. & Longo, D. L. Precision medicine — personalized, problematic, and promising. N. Engl. J. Med. 372, 2229–2234 (2015). This article discusses the overall scope and potential applications of precision medicine, and analyses questions regarding its implementation in the practice of medicine.
Chan, A. C. & Behrens, T. W. Personalizing medicine for autoimmune and inflammatory diseases. Nat. Immunol. 14, 106–109 (2013).
Cox, D. B., Platt, R. J. & Zhang, F. Therapeutic genome editing: prospects and challenges. Nat. Med. 21, 121–131 (2015).
Author information
Authors and Affiliations
Contributions
All authors contributed equally to all aspects of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Supplementary information
Supplementary information S1 (figure)
Screenshot of CellFateScout result. (PDF 613 kb)
Supplementary information S2 (figure)
Flow diagram for utilization of human IBD clinical biosamples for integrated –omics analyses. (PDF 439 kb)
Rights and permissions
About this article
Cite this article
de Souza, H., Fiocchi, C. & Iliopoulos, D. The IBD interactome: an integrated view of aetiology, pathogenesis and therapy. Nat Rev Gastroenterol Hepatol 14, 739–749 (2017). https://doi.org/10.1038/nrgastro.2017.110
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrgastro.2017.110
This article is cited by
-
DHX9 maintains epithelial homeostasis by restraining R-loop-mediated genomic instability in intestinal stem cells
Nature Communications (2024)
-
Oral pyroptosis nanoinhibitor for the treatment of inflammatory bowel disease
Nano Research (2024)
-
Inhibition of GSDMD-mediated pyroptosis triggered by Trichinella spiralis intervention contributes to the alleviation of DSS-induced ulcerative colitis in mice
Parasites & Vectors (2023)
-
Inflammatory bowel disease biomarkers revealed by the human gut microbiome network
Scientific Reports (2023)
-
Experimental evidence of shikonin as a novel intervention for anti-inflammatory effects
Naunyn-Schmiedeberg's Archives of Pharmacology (2023)