Key Points
-
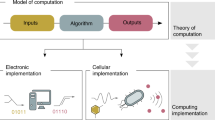
The notion of computation is none other than a systematic way of processing information, and thus computation is central to the function of biological systems, as it is crucial for complex man-made machinery.
-
Whereas biological computing is ubiquitous in living systems, the capacity to engineer new biological computing systems will open the way to an unprecedented level of rational control over living matter that can be used in all areas of biological engineering and medicine
-
Current engineering effort is split between biochemical systems that function in carefully constituted settings and biological systems that operate in living cells or cell ensembles. The two approaches are complementary because biochemical systems show what is possible in principle, whereas biological systems must deal with the complexity of the host and thus are at this point simpler and smaller in scale.
-
The construction of molecular computing systems has been inspired by known theoretical models of computation, such as state machines and logic and analogue circuits. Each model is best suited for a different class of tasks.
-
The logic circuits model has spawned a large number of implementations both in the test tube and in living cells, with the basic building blocks comprising DNA oligomers in the test tube and re-engineered regulatory switches in living cells. Recent achievements include neural-like network with associative memory made of DNA switches, a trainable ribozyme-based molecular network, a number of distributed logic gates in bacteria and yeast and a cell-type classifier for cancer cell detection and destruction.
-
Molecular systems inspired by state machines were implemented with both biochemical and biological approaches, resulting in molecular finite automaton and recombinase-based counter.
Abstract
The task of information processing, or computation, can be performed by natural and man-made 'devices'. Man-made computers are made from silicon chips, whereas natural 'computers', such as the brain, use cells and molecules. Computation also occurs on a much smaller scale in regulatory and signalling pathways in individual cells and even within single biomolecules. Indeed, much of what we recognize as life results from the remarkable capacity of biological building blocks to compute in highly sophisticated ways. Rational design and engineering of biological computing systems can greatly enhance our ability to study and to control biological systems. Potential applications include tissue engineering and regeneration and medical treatments. This Review introduces key concepts and discusses recent progress that has been made in biomolecular computing.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Wiener, N. Cybernetics (MIT Press, 1948).
Hartwell, L. H., Hopfield, J. J., Leibler, S. & Murray, A. W. From molecular to modular cell biology. Nature 402, C47–C52 (1999).
Nurse, P. Life, logic and information. Nature 454, 424–426 (2008).
Keasling, J. D. Manufacturing molecules through metabolic engineering. Science 330, 1355–1358 (2010).
Morelli, A. E. & Thomson, A. W. Tolerogenic dendritic cells and the quest for transplant tolerance. Nature Rev. Immunol. 7, 610–621 (2007).
Khademhosseini, A., Langer, R., Borenstein, J. & Vacanti, J. P. Microscale technologies for tissue engineering and biology. Proc. Natl Acad. Sci. USA 103, 2480–2487 (2006).
Bockamp, E. et al. Conditional transgenic mouse models: from the basics to genome-wide sets of knockouts and current studies of tissue regeneration. Regen. Med. 3, 217–235 (2008).
Kartsson, M., Weber, W. & Fussenegger, M. in Methods in Enzymology: Synthetic Biology, Part A. Methods for Part/Device Characterization and Chassis Engineering Vol. 497 (ed. Voigt, C.) 239–253 (2011).
Sugita, M. Functional analysis of chemical systems in vivo using a logical circuit equivalent. J. Theor. Biol. 1, 415–430 (1961).
Adleman, L. M. Molecular computation of solutions to combinatorial problems. Science 266, 1021–1024 (1994). This paper provides the first experimentally shown computation with DNA molecules.
Lipton, R. J. DNA solution of hard computational problems. Science 268, 542–545 (1995).
Faulhammer, D., Cukras, A. R., Lipton, R. J. & Landweber, L. F. Molecular computation: RNA solutions to chess problems. Proc. Natl Acad. Sci. USA 97, 1385–1389 (2000).
Knight, T. F. & Sussman, G. J. in Unconventional Models of Computation (eds Calude, C.S., Casti, J. & Dinneen, M.J.) 257–272 (Springer, 1998).
Benner, S. A. & Sismour, A. M. Synthetic biology. Nature Rev. Genet. 6, 533–543 (2005).
Baker, D. et al. Engineering life: building a fab for biology. Sci. Am. 294, 44–51 (2006).
Tan, C. M., Song, H., Niemi, J. & You, L. C. A synthetic biology challenge: making cells compute. Mol. Biosyst. 3, 343–353 (2007).
Xie, Z., Wroblewska, L., Prochazka, L., Weiss, R. & Benenson, Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science 333, 1307–1311 (2011). This paper describes a large-scale control system in human cells that identifies a cancerous cell state on the basis of multiple endogenous miRNA markers and produces a fluorescent or apoptosis-inducing protein following positive detection.
Shapiro, E. & Benenson, Y. Bringing DNA computers to life. Sci. Am. 294, 44–51 (2006).
Voigt, C. A. & Keasling, J. D. Programming cellular function. Nature Chem. Biol. 1, 304–307 (2005).
Nelson, V. P., Nagle, H. T., Irwin, J. D. & Carroll, B. D. Digital Logic Circuit Analysis and Design (Prentice Hall, 1995).
Monod, J. & Jacob, F. General conclusions — teleonomic mechanisms in cellular metabolism, growth, and differentiation. Cold Spring Harb. Symp. Quant. Biol. 26, 389–401 (1961).
Sugita, M. Functional analysis of chemical systems in vivo using a logical circuit equivalent. V. Molecular biological interpretation of self-reproducing automata theory and chemico-physical interpretation of information in biological systems. J. Theor. Biol. 53, 223–237 (1975).
Glass, L. & Kauffman, S. A. Logical analysis of continuous, nonlinear biochemical control networks. J. Theor. Biol. 39, 103–129 (1973).
Kauffman, S., Peterson, C., Samuelsson, B. & Troein, C. Random Boolean network models and the yeast transcriptional network. Proc. Natl Acad. Sci. USA 100, 14796–14799 (2003).
Ptashne, M. Principles of a switch. Nature Chem. Biol. 7, 484–487 (2011).
Gardner, T. S., Cantor, C. R. & Collins, J. J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000).
Ajo-Franklin, C. M. et al. Rational design of memory in eukaryotic cells. Genes Dev. 21, 2271–2276 (2007).
Sugita, M. Functional analysis of chemical systems in vivo using a logical circuit equivalent. II. The idea of a molecular automaton. J. Theor. Biol. 4, 179–184 (1963).
Arkin, A. & Ross, J. Computational functions in biochemical reaction networks. Biophys. J. 67, 560–578 (1994).
Weiss, R., Homsy, G. E. & Knight, T. F. in Evolution as Computation: DIMACS Workshop (eds Landweber, L.F. & Winfree, E.) 275–295 (Springer, 1999).
Tamsir, A., Tabor, J. J. & Voigt, C. A. Robust multicellular computing using genetically encoded NOR gates and chemical 'wires'. Nature 469, 212–215 (2011). An example of complex, distributed logic circuit that uses universal transcription-based NOR gates is provided in this paper.
Buchler, N. E., Gerland, U. & Hwa, T. On schemes of combinatorial transcription logic. Proc. Natl Acad. Sci. USA 100, 5136–5141 (2003).
Benenson, Y. RNA-based computation in live cells. Curr. Opin. Biotechnol. 20, 471–478 (2009).
Basu, S., Gerchman, Y., Collins, C. H., Arnold, F. H. & Weiss, R. A synthetic multicellular system for programmed pattern formation. Nature 434, 1130–1134 (2005).
Regot, S. et al. Distributed biological computation with multicellular engineered networks. Nature 469, 207–211 (2011). This study describe a flexible platform for distributed computation with yeast strains based on mating pathway.
Korn, G. A. & Korn, T. M. Electronic Analog and Hybrid Computers (McGraw–Hill, 1964).
Oishi, K. & Klavins, E. Biomolecular implementation of linear I/O systems. IET Syst. Biol. 5, 252–260 (2011).
McCulloch, W. S. & Pitts, W. A logical calculus of the ideas immanent in nervous activity. Bull. Math. Biol. 5, 115–133 (1943).
Sugita, M. & Fukuda, N. Functional analysis of chemical systems in vivo using a logical circuit equivalent. III. Analysis using a digital circuit combined with an analogue computer. J. Theor. Biol. 5, 412–418 (1963).
Hjelmfelt, A., Weinberger, E. D. & Ross, J. Chemical implementation of neural networks and Turing machines. Proc. Natl Acad. Sci. USA 88, 10983–10987 (1991).
Qian, L. L., Winfree, E. & Bruck, J. Neural network computation with DNA strand displacement cascades. Nature 475, 368–372 (2011). A DNA circuit implementing a small neural network with associative memory properties is described in this paper.
Stahl, W. R. & Goheen, H. E. Molecular algorithms. J. Theor. Biol. 5, 266–287 (1963).
Baer, R. M. & Martinez, H. M. Automata and biology. Annu. Rev. Biophys. Bioeng. 3, 255–291 (1974).
Benenson, Y. et al. Programmable and autonomous computing machine made of biomolecules. Nature 414, 430–434 (2001). This was the first experimental implementation of a finite automaton model of computation with molecular building blocks.
Bennett, C.H. The thermodynamics of computation — a review. Int. J. Theor. Phys. 21, 905–940 (1982).
Shapiro, E. & Karunaratne, K. S. G. Method and system of computing similar to a Turing machine. US Patent 6266569 (2001).
Rothemund, P. W. K. in DNA-Based Computers. Proceedings of a DIMACS Workshop (eds Lipton, R.J. & Baum, E.B.) 75–120 (1995).
Ham, T. S., Lee, S. K., Keasling, J. D. & Arkin, A. P. Design and construction of a double inversion recombination switch for heritable sequential genetic memory. PLoS ONE 3 (2008).
Head, T. Formal language theory and DNA — an analysis of the generative capacity of specific recombinant behaviors. Bull. Math. Biol. 49, 737–759 (1987).
Paun, G. & Rozenberg, G. A guide to membrane computing. Theor. Comput. Sci. 287, 73–100 (2002).
Adamatzky, A. Universal dynamical computation in multidimensional excitable lattices. Int. J. Theor. Phys. 37, 3069–3108 (1998).
Harju, T., Petre, I., Rogojin, V. & Rozenberg, G. Patterns of simple gene assembly in ciliates. Discrete Appl. Math. 156, 2581–2597 (2008).
Nowacki, M. et al. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature 451, 153–158 (2008).
Cardelli, L. in Algorithmic Bioprocesses (eds Condon, A., Harel, D., Kok, J.N., Salomaa, A. & Winfree, E.) 429–462 (Springer, 2009).
Abelson, H. et al. Amorphous computing. Commun. ACM 43, 74–82 (2000).
Rackham, O. & Chin, J. W. Cellular logic with orthogonal ribosomes. J. Am. Chem. Soc. 127, 17584–17585 (2005).
Desilva, A. P., Gunaratne, H. Q. N. & McCoy, C. P. A molecular photoionic AND gate based on fluorescent signalling. Nature 364, 42–44 (1993).
Macdonald, J. et al. Medium scale integration of molecular logic gates in an automaton. Nano Lett. 6, 2598–2603 (2006).
Pei, R. J., Matamoros, E., Liu, M. H., Stefanovic, D. & Stojanovic, M. N. Training a molecular automaton to play a game. Nature Nanotechnol. 5, 773–777 (2010). This paper describes an advanced DNAzyme-based circuitry that can be programmed to perform different tasks according to molecular 'training instructions' it has received.
Elbaz, J. et al. DNA computing circuits using libraries of DNAzyme subunits. Nature Nanotechnol. 5, 417–422 (2010).
Yin, P., Choi, H. M. T., Calvert, C. R. & Pierce, N. A. Programming biomolecular self-assembly pathways. Nature 451, 318–322 (2008).
Zhang, D. Y., Turberfield, A. J., Yurke, B. & Winfree, E. Engineering entropy-driven reactions and networks catalyzed by DNA. Science 318, 1121–1125 (2007).
Seelig, G., Soloveichik, D., Zhang, D. Y. & Winfree, E. Enzyme-free nucleic acid logic circuits. Science 314, 1585–1588 (2006).
Qian, L. L. & Winfree, E. Scaling up digital circuit computation with DNA strand displacement cascades. Science 332, 1196–1201 (2011). In this paper, a large-scale DNA circuit that is based on strand displacement reactions is described.
Lewandoski, M. Conditional control of gene expression in the mouse. Nature Rev. Genet. 2, 743–755 (2001).
Lambowitz, A. M. & Zimmerly, S. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb. Perspect. Biol. 3, a003616 (2011).
Jaenisch, R. & Bird, A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genet. 33, 245–254 (2003).
Haynes, K. A. & Silver, P. A. Synthetic reversal of epigenetic silencing. J. Biol. Chem. 286, 27176–27182 (2011).
Davidson, E. A. & Ellington, A. D. Synthetic RNA circuits. Nature Chem. Biol. 3, 23–28 (2007).
Isaacs, F. J., Dwyer, D. J. & Collins, J. J. RNA synthetic biology. Nature Biotech. 24, 545–554 (2006).
Win, M. N. & Smolke, C. D. Higher-order cellular information processing with synthetic RNA devices. Science 322, 456–460 (2008).
Lucks, J. B., Qi, L., Mutalik, V. K., Wang, D. & Arkin, A. P. Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proc. Natl Acad. Sci. USA 108, 8617–8622 (2011). This study described and tested, in the engineering context, an antisense RNA-based regulatory mechanism in bacteria.
Culler, S. J., Hoff, K. G. & Smolke, C. D. Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science 330, 1251–1255 (2010).
Rinaudo, K. et al. A universal RNAi-based logic evaluator that operates in mammalian cells. Nature Biotech. 25, 795–801 (2007).
Leisner, M., Bleris, L., Lohmueller, J., Xie, Z. & Benenson, Y. Rationally designed logic integration of regulatory signals in mammalian cells. Nature Nanotechnol. 5, 666–670 (2010).
Tu, K. C. & Bassler, B. L. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 21, 221–233 (2007).
Levine, E., Zhang, Z., Kuhlman, T. & Hwa, T. Quantitative characteristics of gene regulation by small RNA. PLoS Biol. 5, 1998–2010 (2007).
Niazov, T., Baron, R., Katz, E., Lioubashevski, O. & Willner, I. Concatenated logic gates using four coupled biocatalysts operating in series. Proc. Natl. Acad. Sci. USA 103, 17160–17163 (2006).
Zhou, J., Arugula, M. A., Halamek, J., Pita, M. & Katz, E. Enzyme-based NAND and NOR logic gates with modular design. J. Phys. Chem. B 113, 16065–16070 (2009).
Privman, V., Strack, G., Solenov, D., Pita, M. & Katz, E. Optimization of enzymatic biochemical logic for noise reduction and scalability: how many biocomputing gates can be interconnected in a circuit? J. Phys. Chem. B 112, 11777–11784 (2008).
Wagner, N., Alesebi, S. & Ashkenasy, G. How symmetry and order affect logic operations and computation in catalytic chemical networks. J. Comput. Theor. Nanosci. 8, 471–480 (2011).
Brent, R. & Ptashne, M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell 43, 729–736 (1985).
Kramer, B. & Fussenegger, M. in Encyclopedia of Genetics, Genomics, Proteomics and Bioinformatics (Wiley, 2005).
Kramer, B. P., Fischer, C. & Fussenegger, M. BioLogic gates enable logical transcription control in mammalian cells. Biotechnol. Bioeng. 87, 478–484 (2004). This work demonstrated a three-input logic gate on a mammalian promoter.
Cox, R. S., Surette, M. G. & Elowitz, M. B. Programming gene expression with combinatorial promoters. Mol. Syst. Biol. 3, 145 (2007).
Guet, C. C., Elowitz, M. B., Hsing, W. H. & Leibler, S. Combinatorial synthesis of genetic networks. Science 296, 1466–1470 (2002).
Hooshangi, S., Thiberge, S. & Weiss, R. Ultrasensitivity and noise propagation in a synthetic transcriptional cascade. Proc. Natl Acad. Sci. USA 102, 3581–3586 (2005).
Ellis, T., Wang, X. & Collins, J. J. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nature Biotech. 27, 465–471 (2009).
Tabor, J. J. et al. A synthetic genetic edge detection program. Cell 137, 1272–1281 (2009).
Wang, B. J., Kitney, R. I., Joly, N. & Buck, M. Engineering modular and orthogonal genetic logic gates for robust digital-like synthetic biology. Nature Commun. 2, 18 Oct 2011 (doi: 10.1038/ncomms1516).
Ye, H. F., Daoud- El Baba, M., Peng, R. W. & Fussenegger, M. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science 332, 1565–1568 (2011).
Nissim, L. & Bar-Ziv, R. H. A tunable dual-promoter integrator for targeting of cancer cells. Mol. Syst. Biol. 6, 444 (2010).
Grunberg, R. & Serrano, L. Strategies for protein synthetic biology. Nucleic Acids Res. 38, 2663–2675 (2010).
Park, S. H., Zarrinpar, A. & Lim, W. A. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science 299, 1061–1064 (2003).
Skerker, J. M. et al. Rewiring the specificity of two-component signal transduction systems. Cell 133, 1043–1054 (2008).
Turner, B. M. Cellular memory and the histone code. Cell 111, 285–291 (2002).
Appella, E. & Anderson, C. W. Signaling to p53: breaking the posttranslational modification code. Pathol. Biol. 48, 227–245 (2000).
Dueber, J. E., Yeh, B. J., Chak, K. & Lim, W. A. Reprogramming control of an allosteric signaling switch through modular recombination. Science 301, 1904–1908 (2003).
Grilly, C., Stricker, J., Pang, W. L., Bennett, M. R. & Hasty, J. A synthetic gene network for tuning protein degradation in Saccharomyces cerevisiae. Mol. Syst. Biol. 3, 127 (2007).
Wang, H. Proving theorems by pattern recognition I. Commun. ACM 40, 1–42 (1961).
Robinson, R. M. Undecidability and non-periodicity for tilings of plane. Invent. Math. 12, 177–209 (1971).
Winfree, E. Algorithmic Self-assembly of DNA. Thesis, California Institute of Technology (1998).
Winfree, E., Liu, F. R., Wenzler, L. A. & Seeman, N. C. Design and self-assembly of two-dimensional DNA crystals. Nature 394, 539–544 (1998).
Mao, C. D., LaBean, T. H., Reif, J. H. & Seeman, N. C. Logical computation using algorithmic self-assembly of DNA triple-crossover molecules. Nature 407, 493–496 (2000).
Schulman, R. & Winfree, E. Synthesis of crystals with a programmable kinetic barrier to nucleation. Proc. Natl Acad. Sci. USA 104, 15236–15241 (2007).
Barish, R. D., Schulman, R., Rothemund, P. W. K. & Winfree, E. An information-bearing seed for nucleating algorithmic self-assembly. Proc. Natl Acad. Sci. USA 106, 6054–6059 (2009). This is the most complex DNA-tiling computation to date, implementing a counter to 17.
Chen, J. H. & Seeman, N. C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature 350, 631–633 (1991).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Douglas, S. M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Lund, K. et al. Molecular robots guided by prescriptive landscapes. Nature 465, 206–210 (2010).
Delebecque, C. J., Lindner, A. B., Silver, P. A. & Aldaye, F. A. Organization of intracellular reactions with rationally designed RNA assemblies. Science 333, 470–474 (2011). This paper is an example of nucleic acids nanotechnology use in living cells for improved efficiency of a bioproduction pathway.
Douglas, S. M., Bachelet, I. & Church, G. M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 335, 831–834 (2012). This study combines DNA nanotechnology with the logic gate concept for selective cell targeting.
Smith, W. D. in DNA-Based Computers. Proceedings of a DIMACS Workshop (eds Lipton, R.J. & Baum, E.B.) 121–186 (American Mathematical Society, 1995).
Sakamoto, K. et al. State transitions by molecules. Biosystems 52, 81–91 (1999).
Benenson, Y., Adar, R., Paz-Elizur, T., Livneh, Z. & Shapiro, E. DNA molecule provides a computing machine with both data and fuel. Proc. Natl Acad. Sci. USA 100, 2191–2196 (2003).
Soreni, M., Yogev, S., Kossoy, E., Shoham, Y. & Keinan, E. Parallel biomolecular computation on surfaces with advanced finite automata. J. Am. Chem. Soc. 127, 3935–3943 (2005).
Adar, R. et al. Stochastic computing with biomolecular automata. Proc. Natl Acad. Sci. USA 101, 9960–9965 (2004).
Benenson, Y., Gil, B., Ben-Dor, U., Adar, R. & Shapiro, E. An autonomous molecular computer for logical control of gene expression. Nature 429, 423–429 (2004). This paper shows the first biochemical prototype of a large-scale control circuit combining sophisticated sensing, computation and actuation modules to make decisions based on multiple environmental cues.
Gil, B., Kahan-Hanum, M., Skirtenko, N., Adar, R. & Shapiro, E. Detection of multiple disease indicators by an autonomous biomolecular computer. Nano Lett. 11, 2989–2996 (2011).
Ran, T., Kaplan, S. & Shapiro, E. Molecular implementation of simple logic programs. Nature Nanotechnol. 4, 642–648 (2009).
Friedland, A. E. et al. Synthetic gene networks that count. Science 324, 1199–1202 (2009). An impressive example of recombinase-based circuitry is discussed in this paper that can irreversibly record up to three consecutive events.
Wang, Z. G., Elbaz, J., Remacle, F., Levine, R. D. & Willner, I. All-DNA finite-state automata with finite memory. Proc. Natl Acad. Sci. USA 107, 21996–22001 (2010).
Dayarian, A., Chaves, M., Sontag, E. D. & Sengupta, A. M. Shape, size, and robustness: feasible regions in the parameter space of biochemical networks. PLoS Comput. Biol. 5, e1000256 (2009).
Kim, H. D. & O'Shea, E. K. A quantitative model of transcription factor-activated gene expression. Nature Struct. Mol. Biol. 15, 1192–1198 (2008).
Rosenfeld, N., Young, J. W., Alon, U., Swain, P. S. & Elowitz, M. B. Gene regulation at the single-cell level. Science 307, 1962–1965 (2005).
Gregor, T., Tank, D. W., Wieschaus, E. F. & Bialek, W. Probing the limits to positional information. Cell 130, 153–164 (2007).
Scott, M., Hwa, T. & Ingalls, B. Deterministic characterization of stochastic genetic circuits. Proc. Natl Acad. Sci. USA 104, 7402–7407 (2007).
Ando, H., Sinha, S., Storni, R. & Aihara, K. Synthetic gene networks as potential flexible parallel logic gates. Europhys. Lett. 93, 50001 (2011).
Klavins, E. in Proc. 49th IEEE Conf. Decision Control 2547–2553 (2010).
Bleris, L. et al. Synthetic incoherent feedforward circuits show adaptation to the amount of their genetic template. Mol. Syst. Biol. 7, 519 (2011).
Elowitz, M. B. & Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000).
Tigges, M., Marquez-Lago, T. T., Stelling, J. & Fussenegger, M. A tunable synthetic mammalian oscillator. Nature 457, 309–312 (2009).
Stricker, J. et al. A fast, robust and tunable synthetic gene oscillator. Nature 456, 516–519 (2008).
Becskei, A. & Serrano, L. Engineering stability in gene networks by autoregulation. Nature 405, 590–593 (2000).
Marchisio, M. A. & Stelling, J. Computational design of synthetic gene circuits with composable parts. Bioinformatics 24, 1903–1910 (2008).
Beal, J., Lu, T. & Weiss, R. Automatic compilation from high-level biologically-oriented programming language to genetic regulatory networks. PLoS ONE 6, e22490 (2011).
Canton, B., Labno, A. & Endy, D. Refinement and standardization of synthetic biological parts and devices. Nature Biotech. 26, 787–793 (2008).
Turing, A. M. On computable numbers, with an application to the Entscheidungsproblem. Proc. Lond. Math. Soc. 42, 230–265 (1937).
Acknowledgements
The author's research is funded by ETH Zurich, a US National Institutes of Health and National Cancer Institute grant (5R01CA155320) and a European Research Council starting grant. He wishes to thank F. Rudolf for discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author holds a pending patent application covering some of his work discussed in this Review.
Supplementary information
Supplementary information S1 (figure)
Tiling implementation of Turing machine computation. (PDF 164 kb)
Related links
Glossary
- Input
-
A unit of information that is processed by a computing system, or a collection of all such units.
- Output
-
A unit of information that is produced as a result of computation, or a collection of all such units.
- Mapping
-
A specific relationship between the inputs and the outputs of computation expressed as a mathematical function or as a computation procedure (program); it can be formally described as a collection of all pairs ([inputs], [outputs]), where [inputs] is a specific combination of legitimate inputs, and [outputs] is a result of computation for this combination.
- Models of computation
-
Specific approaches towards implementing information-processing tasks; they are usually required to be universal.
- Molecular computer
-
A design framework that enables construction of molecular systems that are capable of implementing desired input–output mappings between molecular inputs and outputs or a specific implementation of such a system.
- Autonomous systems
-
A molecular computer that does not require external interference apart from initializing the computer components and (optionally) the inputs.
- Gene circuit
-
A set of engineered genes that can be implanted into a living cell and, following their expression, can form functional biological networks comprising these genes and their products (RNA and protein).
- Logic functions
-
Mappings between multiple inputs and a single output, where both the inputs and the output can only take values of zero and one (or false and true).
- Logic circuits
-
Specific arrangements of logic gates that can compute specific logic functions.
- Universal set
-
A collection of gate types that can be used to compute any logic function.
- Universal gates
-
Gates of a single type that can be used to implement any conceivable logic function.
- Normal form
-
A standard way of expressing logic functions that can be used to represent any logic function.
- Analogue circuits
-
Arrangements of gates that compute continuous-value functions, such as multiplication.
- State machines
-
A class of models of computation that comprise a tape of symbols as data storage and a controller that scans the tape, reads and writes symbols and modifies its own state based on specific transition rules.
- Finite automata
-
A class of state machines that process strings from left to right. The controller scans symbols one by one, changing the state at each step, depending on the current state, according to the rule <current state>, <current symbol> <next state> (and move to the next symbol).
- Belousov–Zhabotinsky reactions
-
A set of coupled chemical processes that do not reach equilibrium for extended periods of time and instead exhibit oscillations or other dynamic features
- Distributed computing
-
A computer architecture that uses multiple stand-alone computing units that interact with each other to accomplish a common computational task.
- Amorphous computing
-
An extreme case of distributed computing with very large number of simple computing units that can move in space and only interact locally.
- McCulloch–Pitts neuron
-
An abstract gate embodying some features of neuron cells, which calculates a weighted sum of the inputs and generates an output of one when this sum is above a certain threshold.
- Logic gate
-
A small computational unit that implements a fixed logic function such as AND between one or two, but sometimes more, inputs.
- Strand displacement
-
A chemical process whereby a single-stranded DNA oligonucleotide replaces the shorter of two strands in a partially double-stranded DNA duplex. This starts with the oligonucleotide binding to the single-stranded section (the 'toehold') and goes to completion because the new duplex has a higher thermodynamic stability.
- Hopfield neural network
-
A class of artificial neural networks in which the individual 'neurons' mutually excite and inhibit each other. The network can be trained with specific input sets (patterns) such that each pattern corresponds to a steady state. After the network has been trained, any new input pattern will cause the network to convert to the state that is closest to this pattern, implementing memory by association.
- Open-loop processes
-
Physical processes or computations whose outputs do not affect the inputs.
Rights and permissions
About this article
Cite this article
Benenson, Y. Biomolecular computing systems: principles, progress and potential. Nat Rev Genet 13, 455–468 (2012). https://doi.org/10.1038/nrg3197
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3197
This article is cited by
-
Non-complementary strand commutation as a fundamental alternative for information processing by DNA and gene regulation
Nature Chemistry (2023)
-
DNA-based programmable gate arrays for general-purpose DNA computing
Nature (2023)
-
Teaching computing for complex problems in civil engineering and geosciences using big data and machine learning: synergizing four different computing paradigms and four different management domains
Journal of Big Data (2023)
-
A tape-reading molecular ratchet
Nature (2022)
-
Chemical-to-mechanical molecular computation using DNA-based motors with onboard logic
Nature Nanotechnology (2022)