Abstract
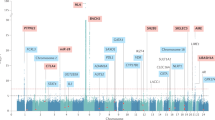
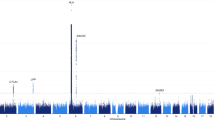
Many endocrine diseases, including type 1 diabetes mellitus, Graves disease, Addison disease and Hashimoto disease, originate as an autoimmune reaction that affects disease-specific target organs. These autoimmune diseases are characterized by the development of specific autoantibodies and by the presence of autoreactive T cells. They are caused by a complex genetic predisposition that is attributable to multiple genetic variants, each with a moderate-to-low effect size. Most of the genetic variants associated with a particular autoimmune endocrine disease are shared between other systemic and organ-specific autoimmune and inflammatory diseases, such as rheumatoid arthritis, coeliac disease, systemic lupus erythematosus and psoriasis. Here, we review the shared and specific genetic background of autoimmune diseases, summarize their treatment options and discuss how identifying the genetic and environmental factors that predispose patients to an autoimmune disease can help in the diagnosis and monitoring of patients, as well as the design of new treatments.
Key Points
-
Autoimmune diseases share a large proportion of their genetic background; shared genes and pathways might serve as common drug targets across diseases
-
Genetic studies are discovering new pathways relevant to autoimmune diseases
-
Determining genetic profiles for common autoimmune diseases could improve understanding of the underlying pathways and help identify novel drug targets for less common and rare autoimmune diseases
-
Genetic profiling will help identify individuals at risk of autoimmune diseases, which is important for timely diagnosis and treatment
-
Genetic, microRNA, metabolomic, microbiome and autoimmune profiles can serve as biomarkers for monitoring disease progression and response to treatment
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Qiao, S.-W., Iversen, R., Ráki, M. & Sollid, L. M. The adaptive immune response in celiac disease. Semin. Immunopathol. 34, 523–540 (2012).
Van Lummel, M., Zaldumbide, A. & Roep, B. O. Changing faces, unmasking the β-cell: post-translational modification of antigens in type 1 diabetes. Curr. Opin. Endocrinol. Diabetes Obes. 20, 299–306 (2013).
Weetman, A. P. Cellular immune responses in autoimmune thyroid disease. Clin. Endocrinol. 61, 405–413 (2004).
Richard-Miceli, C. & Criswell, L. A. Emerging patterns of genetic overlap across autoimmune disorders. Genome Med. 4, 6 (2012).
Zhernakova, A., Van Diemen, C. C. & Wijmenga, C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat. Rev. Genet. 10, 43–55 (2009).
Solovieff, N., Cotsapas, C., Lee, P. H., Purcell, S. M. & Smoller, J. W. Pleiotropy in complex traits: challenges and strategies. Nat. Rev. Genet. 14, 483–495 (2013).
Cooper, J. D. et al. Seven newly identified loci for autoimmune thyroid disease. Hum. Mol. Genet. 21, 5202–5208 (2012).
Trynka, G. et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Genet. 43, 1193–1201 (2011).
Tsoi, L. C. et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet. 44, 1341–1348 (2012).
Eyre, S. et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat. Genet. 44, 1336–1340 (2012).
Zhernakova, A. et al. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet. 7, e1002004 (2011).
Smyth, D. J. et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N. Engl. J. Med. 359, 2767–2777 (2008).
Hsu, L. N. & Armstrong, A. W. Psoriasis and autoimmune disorders: a review of the literature. J. Am. Acad. Dermatol. 67, 1076–1079 (2012).
Hemminki, K., Li, X., Sundquist, K. & Sundquist, J. Shared familial aggregation of susceptibility to autoimmune diseases. Arthritis Rheum. 60, 2845–2847 (2009).
Skinningsrud, B. et al. Multiple loci in the HLA complex are associated with Addison's disease. J. Clin. Endocrinol. Metab. 96, E1703–E1708 (2011).
Zurawek, M. et al. A coding variant in NLRP1 is associated with autoimmune Addison's disease. Hum. Immunol. 71, 530–534 (2010).
Magitta, N. F. et al. A coding polymorphism in NALP1 confers risk for autoimmune Addison's disease and type 1 diabetes. Genes Immun. 10, 120–124 (2009).
Skinningsrud, B. et al. Mutation screening of PTPN22: association of the 1858T-allele with Addison's disease. Eur. J. Hum. Genet. 16, 977–982 (2008).
Criswell, L. A. et al. Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am. J. Hum. Genet. 76, 561–571 (2005).
Vaidya, B. et al. Association analysis of the cytotoxic T lymphocyte antigen-4 (CTLA-4) and autoimmune regulator-1 (AIRE-1) genes in sporadic autoimmune Addison's disease. J. Clin. Endocrinol. Metab. 85, 688–691 (2000).
Brozzetti, A. et al. Cytotoxic T lymphocyte antigen-4 Ala17 polymorphism is a genetic marker of autoimmune adrenal insufficiency: Italian association study and meta-analysis of European studies. Eur. J. Endocrinol. 162, 361–369 (2010).
Owen, C. J. et al. Analysis of the Fc receptor-like-3 (FCRL3) locus in Caucasians with autoimmune disorders suggests a complex pattern of disease association. J. Clin. Endocrinol. Metab. 92, 1106–1111 (2007).
Skinningsrud, B. et al. Polymorphisms in CLEC16A and CIITA at 16p13 are associated with primary adrenal insufficiency. J. Clin. Endocrinol. Metab. 93, 3310–3317 (2008).
Husebye, E. S. & Anderson, M. S. Review autoimmune polyendocrine syndromes: clues to type 1 diabetes pathogenesis. Immunity 32, 479–487 (2010).
Gan, E. H., Mitchell, A. L., Macarthur, K. & Pearce, S. H. S. The role of a nonsynonymous CD226 (DNAX-accessory molecule-1) variant (Gly 307Ser) in isolated Addison's disease and autoimmune polyendocrinopathy type 2 pathogenesis. Clin. Endocrinol. 75, 165–168 (2011).
Song, G., Bae, S.-C., Choi, S., Ji, J. & Lee, Y. Association between the CD226 rs763361 polymorphism and susceptibility to autoimmune diseases: a meta-analysis. Lupus 21, 1522–1530 (2012).
Plagnol, V. et al. Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet. 7, e1002216 (2011).
Todd, J. A. et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 39, 857–864 (2008).
Jostins, L. et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012).
Gallo, V. et al. Alterations of the autoimmune regulator transcription factor and failure of central tolerance: APECED as a model. Expert. Rev. Clin. Immunol. 9, 43–51 (2013).
Pearce, S. H. et al. A common and recurrent 13-bp deletion in the autoimmune regulator gene in British kindreds with autoimmune polyendocrinopathy type 1. Am. J. Hum. Genet. 63, 1675–1684 (1998).
Terao, C. et al. The human AIRE gene at chromosome 21q22 is a genetic determinant for the predisposition to rheumatoid arthritis in Japanese population. Hum. Mol. Genet. 20, 2680–2685 (2011).
Barrett, J. C. et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 41, 703–707 (2009).
Stahl, E. A. et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 42, 508–514 (2010).
Bennett, C. L. et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 27, 20–21 (2001).
Dunham, I. et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
McGovern, D. P. B. et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat. Genet. 42, 332–337 (2010).
Nair, R. P. et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-κB pathways. Nat. Genet. 41, 199–204 (2009).
Franke, A. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 42, 1118–1125 (2010).
Ricaño-Ponce, I. & Wijmenga, C. Mapping of immune-mediated disease genes. Annu. Rev. Genomics Hum. Genet. http://dx.doi.org/10.1146/annurev-genom-091212-153450.
Pettigrew, H. D., Teuber, S. S. & Gershwin, M. E. Clinical significance of complement deficiencies. Ann. NY Acad. Sci. 1173, 108–123 (2009).
Pickering, M. C. & Walport, M. J. Links between complement abnormalities and systemic lupus erythematosus. Rheumatology 39, 133–141 (2000).
Lee-Kirsch, M. A. et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat. Genet. 39, 1065–1067 (2007).
Namjou, B. et al. Evaluation of the TREX1 gene in a large multi-ancestral lupus cohort. Genes Immun. 12, 270–279 (2011).
Nejentsev, S., Walker, N., Riches, D., Egholm, M. & Todd, J. A. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science 324, 387–389 (2009).
Betterle, C. & Morlin, L. Autoimmune Addison's disease. Endocr. Dev. 20, 161–172 (2011).
Maahs, D. M., West, N. A., Lawrence, J. M. & Mayer-Davis, E. J. Epidemiology of type 1 diabetes. Endocrinol. Metab. Clin. North Am. 39, 481–497 (2010).
Schön, M. P. & Boehncke, W.-H. Psoriasis. N. Engl. J. Med. 352, 1899–1912 (2005).
Kahaly, G. J. Polyglandular autoimmune syndromes. Eur. J. Endocrinol. 161, 11–20 (2009).
Li, M., Li, C. & Guan, W. Evaluation of coverage variation of SNP chips for genome-wide association studies. Eur. J. Hum. Genet. 16, 635–643 (2008).
Strange, A. et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat. Genet. 42, 985–990 (2010).
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Wacklin, P. et al. The duodenal microbiota composition of adult celiac disease patients is associated with the clinical manifestation of the disease. Inflamm. Bowel Dis. 19, 934–941 (2013).
Ogrendik, M. Rheumatoid arthritis is linked to oral bacteria: etiological association. Mod. Rheumatol. 19, 453–456 (2009).
Fahlén, A., Engstrand, L., Baker, B. S., Powles, A. & Fry, L. Comparison of bacterial microbiota in skin biopsies from normal and psoriatic skin. Arch. Dermatol. Res. 304, 15–22 (2012).
Brown, C. T. et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS ONE 6, e25792 (2011).
Knip, M. & Simell, O. Environmental triggers of type 1 diabetes. Cold Spring Harb. Perspect. Med. 2, a007690 (2012).
Rausch, P. et al. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc. Natl Acad. Sci. USA 108, 19030–19035 (2011).
Markle, J. G. M. et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339, 1084–1088 (2013).
Liu, Y. et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol. 31, 142–147 (2013).
Park, S.-H. et al. Hypermethylation of EBF3 and IRX1 genes in synovial fibroblasts of patients with rheumatoid arthritis. Mol. Cells 35, 298–304 (2013).
Klareskog, L., Catrina, A. I. & Paget, S. Rheumatoid arthritis. Lancet 373, 659–672 (2009).
Kiyohara, C. et al. Risk modification by CYP1A1 and GSTM1 polymorphisms in the association of cigarette smoking and systemic lupus erythematosus in a Japanese population. Scand. J. Rheumatol. 41, 103–109 (2012).
Badenhoop, K. et al. MHC-environment interactions leading to type 1 diabetes: feasibility of an analysis of HLA DR-DQ alleles in relation to manifestation periods and dates of birth. Diabetes Obes. Metab. 11 (Suppl. 1), 88–91 (2009).
Tabas, I. & Glass, C. K. Anti-inflammatory therapy in chronic disease: challenges and opportunities. Science 339, 166–172 (2013).
Scott, D. L., Wolfe, F. & Huizinga, T. W. J. Rheumatoid arthritis. Lancet 376, 1094–1108 (2010).
Faustman, D. & Davis, M. TNF receptor 2 pathway: drug target for autoimmune diseases. Nat. Rev. Drug Discov. 9, 482–493 (2010).
van Schouwenburg, P. A., Rispens, T. & Wolbink, G. J. Immunogenicity of anti-TNF biologic therapies for rheumatoid arthritis. Nat. Rev. Rheumatol. 9, 164–172 (2013).
Fernando, M. M. A. et al. Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS Genet. 4, e1000024 (2008).
Pan, H.-F. et al. Association of TNF-α promoter-308 A/G polymorphism with susceptibility to systemic lupus erythematosus: a meta-analysis. Rheumatol. Int. 32, 2083–2092 (2012).
Li, N. et al. Association of tumour necrosis factor α (TNF-α) polymorphisms with Graves' disease: A meta-analysis. Clin. Biochem. 41, 881–886 (2008).
Shin, H. D., Yang, S. W., Kim, D. H. & Park, Y. Independent association of tumor necrosis factor polymorphism with type 1 diabetes susceptibility. Ann. NY Acad. Sci. 1150, 76–85 (2008).
Science Signaling Database of Cell Signaling. NF-κB pathway [online], (2013).
Ibrahim, I., Owen, S.-A. & Barton, A. Genetics and the impact on treatment protocols in patients with rheumatoid arthritis. Expert Rev. Clin. Immunol. 8, 509–511 (2012).
Wijbrandts, C. A. et al. The clinical response to infliximab in rheumatoid arthritis is in part dependent on pretreatment tumour necrosis factor alpha expression in the synovium. Ann. Rheum. Dis. 67, 1139–1144 (2008).
Gómez-Reino, J. J., Carmona, L., Valverde, V. R., Mola, E. M. & Montero, M. D. Treatment of rheumatoid arthritis with tumor necrosis factor inhibitors may predispose to significant increase in tuberculosis risk: a multicenter active-surveillance report. Arthritis Rheum. 48, 2122–2127 (2003).
Cui, J. et al. Genome-wide association study and gene expression analysis identifies CD84 as a predictor of response to etanercept therapy in rheumatoid arthritis. PLoS Genet. 9, e1003394 (2013).
Gregory, A. P. et al. TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis. Nature 488, 508–511 (2012).
Auphan, N., DiDonato, J. A., Rosette, C., Helmberg, A. & Karin, M. Immunosuppression by glucocorticoids: inhibition of NF-κB activity through induction of IκB synthesis. Science 270, 286–290 (1995).
Scheinman, R. I., Cogswell, P. C., Lofquist, A. K. & Baldwin, A. S. Role of transcriptional activation of IκBα in mediation of immunosuppression by glucocorticoids. Science 270, 283–286 (1995).
Tegeder, I., Pfeilschifter, J. & Geisslinger, G. Cyclooxygenase-independent actions of cyclooxygenase inhibitors. FASEB J. 15, 2057–2072 (2001).
Dasgupta, S. et al. Antineuroinflammatory effect of NF-κB essential modifier-binding domain peptides in the adoptive transfer model of experimental allergic encephalomyelitis. J. Immunol. 173, 1344–1354 (2004).
Sullivan, B. O., Thompson, A. & Thomas, R. NF-κB as a therapeutic target in autoimmune disease. Expert Opin. Ther. Targets 11, 111–122 (2007).
McIntyre, K. W. et al. A highly selective inhibitor of IκB kinase, BMS-345541, blocks both joint inflammation and destruction in collagen-induced arthritis in mice. Arthritis Rheum. 48, 2652–2659 (2003).
Palombella, V. J. et al. Role of the proteasome and NF-κB in streptococcal cell wall-induced polyarthritis. Proc. Natl Acad. Sci. USA 95, 15671–15676 (1998).
Li, G. et al. Human genetics in rheumatoid arthritis guides a high-throughput drug screen of the CD40 signaling pathway. PLoS Genet. 9, e1003487 (2013).
Miossec, P. & Kolls, J. K. Targeting IL-17 and TH17 cells in chronic inflammation. Nat. Rev. Drug Discov. 11, 763–776 (2012).
Watanabe, N. & Nakajima, H. Coinhibitory molecules in autoimmune diseases. Clin. Dev. Immunol. 2012, 269756 (2012).
Chu, X. et al. A genome-wide association study identifies two new risk loci for Graves' disease. Nat. Genet. 43, 897–901 (2011).
Gregersen, P. K. et al. REL, encoding a member of the NF-κB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat. Genet. 41, 820–823 (2009).
Yang, W. et al. Meta-analysis followed by replication identifies loci in or near CDKN1B, TET3, CD80, DRAM1, and ARID5B as associated with systemic lupus erythematosus in Asians. Am. J. Hum. Genet. 92, 41–51 (2013).
Nakamura, M. et al. Genome-wide association study identifies TNFSF15 and POU2AF1 as susceptibility loci for primary biliary cirrhosis in the Japanese population. Am. J. Hum. Genet. 91, 721–728 (2012).
Dubois, P. C. A. et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 42, 295–302 (2010).
Emery, P. et al. Impact of T-cell costimulation modulation in patients with undifferentiated inflammatory arthritis or very early rheumatoid arthritis: a clinical and imaging study of abatacept (the ADJUST trial). Ann. Rheum. Dis. 69, 510–516 (2010).
Orban, T. et al. Co-stimulation modulation with abatacept in patients with recent-onset type 1 diabetes: a randomised, double-blind, placebo-controlled trial. Lancet 378, 412–419 (2011).
Cui, Y., Sheng, Y. & Zhang, X. Genetic susceptibility to SLE: recent progress from GWAS. J. Autoimmun. 41, 25–33 (2013).
Harley, I. T. W., Kaufman, K. M., Langefeld, C. D., Harley, J. B. & Kelly, J. A. Genetic susceptibility to SLE: new insights from fine mapping and genome-wide association studies. Nat. Rev. Genet. 10, 285–290 (2009).
Flesher, D. L. T., Sun, X., Behrens, T. W., Graham, R. R. & Criswell, L. A. Recent advances in the genetics of systemic lupus erythematosus. Expert Rev. Clin. Immunol. 6, 461–479 (2010).
Bezalel, S., Asher, I., Elbirt, D. & Sthoeger, Z. M. Novel biological treatments for systemic lupus erythematosus: current and future modalities. Isr. Med. Assoc. J. 14, 508–514 (2012).
Kozyrev, S. V. et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat. Genet. 40, 211–216 (2008).
Graham, R. R. et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat. Genet. 40, 1059–1061 (2008).
Furie, R. et al. A phase III, randomized, placebo-controlled study of belimumab, a monoclonal antibody that inhibits B lymphocyte stimulator, in patients with systemic lupus erythematosus. Arthritis Rheum. 63, 3918–3930 (2011).
Dall'era, M. & Chakravarty, E. F. Treatment of mild, moderate, and severe lupus erythematosus: focus on new therapies. Curr. Rheumatol. Rep. 13, 308–316 (2011).
Bronson, P. G., Chaivorapol, C., Ortmann, W., Behrens, T. W. & Graham, R. R. The genetics of type I interferon in systemic lupus erythematosus. Curr. Opin. Immunol. 24, 530–537 (2012).
US National Library of Medicine. ClinicalTrials.gov [online], (2013).
US National Library of Medicine. ClinicalTrials.gov [online], (2013).
Burmester, G. R., Panaccione, R., Gordon, K. B., McIlraith, M. J. & Lacerda, A. P. M. Adalimumab: long-term safety in 23 458 patients from global clinical trials in rheumatoid arthritis, juvenile idiopathic arthritis, ankylosing spondylitis, psoriatic arthritis, psoriasis and Crohn's disease. Ann. Rheum. Dis. 72, 517–524 (2013).
Cleynen, I. & Vermeire, S. Paradoxical inflammation induced by anti-TNF agents in patients with IBD. Nat. Rev. Gastroenterol. Hepatol. 9, 496–503 (2012).
Kopf, M., Bachmann, M. F. & Marsland, B. J. Averting inflammation by targeting the cytokine environment. Nat. Rev. Drug Discov. 9, 703–718 (2010).
Sildorf, S. M., Fredheim, S., Svensson, J. & Buschard, K. Remission without insulin therapy on gluten-free diet in a 6-year old boy with type 1 diabetes mellitus. BMJ Case Rep. http://dx.doi.org/10.1136/bcr.02.2012.5878.
Cosnes, J. et al. Incidence of autoimmune diseases in celiac disease: protective effect of the gluten-free diet. Clin. Gastroenterol. Hepatol. 6, 753–758 (2008).
Zhang, T. et al. Host genes related to paneth cells and xenobiotic metabolism are associated with shifts in human ileum-associated microbial composition. PLoS ONE 7, e30044 (2012).
Pozo-Rubio, T. et al. Immune development and intestinal microbiota in celiac disease. Clin. Dev. Immunol. 2012, 654143 (2012).
Murri, M. et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case–control study. BMC Med. 11, 46 (2013).
Kriegel, M. A. et al. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc. Natl Acad. Sci. USA 108, 11548–11553 (2011).
Anderson, J. L., Edney, R. J. & Whelan, K. Systematic review: faecal microbiota transplantation in the management of inflammatory bowel disease. Aliment. Pharmacol. Ther. 36, 503–516 (2012).
Csizmadia, C. G., Mearin, M. L., Von Blomberg, B. M., Brand, R. & Verloove-Vanhorick, S. P. An iceberg of childhood coeliac disease in the Netherlands. Lancet 353, 813–814 (1999).
Arbuckle, M. R. et al. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N. Engl. J. Med. 349, 1526–1533 (2003).
Herold, K. C., Vignali, D. A., Cooke, A. & Bluestone, J. A. Type 1 diabetes: translating mechanistic observations into effective clinical outcomes. Nat. Rev. Immunol. 13, 243–256 (2013).
Rantapää-Dahlqvist, S. et al. Antibodies against cyclic citrullinated peptide and IgA rheumatoid factor predict the development of rheumatoid arthritis. Arthritis Rheum. 48, 2741–2749 (2003).
Nielen, M. M. J. et al. Specific autoantibodies precede the symptoms of rheumatoid arthritis: a study of serial measurements in blood donors. Arthritis Rheum. 50, 380–386 (2004).
Shepshelovich, D. & Shoenfeld, Y. Prediction and prevention of autoimmune diseases: additional aspects of the mosaic of autoimmunity. Lupus 15, 183–190 (2006).
Hutfless, S., Matos, P., Talor, M. V, Caturegli, P. & Rose, N. R. Significance of prediagnostic thyroid antibodies in women with autoimmune thyroid disease. J. Clin. Endocrinol. Metab. 96, E1466–E1471 (2011).
Husby, S. et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J. Pediatr. Gastroenterol. Nutr. 54, 136–160 (2012).
Chatterjee, N. et al. Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nat. Genet. 45, 400–405 (2013).
Pasquinelli, A. E. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 13, 271–282 (2012).
Cortez, M. A. et al. MicroRNAs in body fluids—the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 8, 467–477 (2011).
Weber, J. A. et al. The microRNA spectrum in 12 body fluids. Clin. Chem. 56, 1733–1741 (2010).
Zibert, J. R. et al. MicroRNAs and potential target interactions in psoriasis. J. Dermatol. Sci. 58, 177–185 (2010).
Sonkoly, E. et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS ONE 2, e610 (2007).
Zahm, A. M. et al. Circulating microRNA is a biomarker of pediatric Crohn disease. J. Pediatr. Gastroenterol. Nutr. 53, 26–33 (2011).
Zhou, Q., Souba, W. W., Croce, C. M. & Verne, G. N. MicroRNA-29a regulates intestinal membrane permeability in patients with irritable bowel syndrome. Gut 59, 775–784 (2010).
Liu, G. & Abraham, E. MicroRNAs in immune response and macrophage polarization. Arterioscler. Thromb. Vasc. Biol. 33, 170–177 (2013).
Bernecker, C. et al. MicroRNAs miR-146a1, miR-155_2, and miR-200a1 are regulated in autoimmune thyroid diseases. Thyroid 22, 1294–1295 (2012).
Kumar, V., Wijmenga, C. & Withoff, S. From genome-wide association studies to disease mechanisms: celiac disease as a model for autoimmune diseases. Semin. Immunopathol. 34, 567–580 (2012).
Xia, J., Joyce, C. E., Bowcock, A. M. & Zhang, W. Noncanonical microRNAs and endogenous siRNAs in normal and psoriatic human skin. Hum. Mol. Genet. 22, 737–748 (2013).
Carlsen, A. L. et al. Circulating microRNA expression profiles associated with systemic lupus erythematosus. Arthritis Rheum. 65, 1324–1334 (2013).
Seeger, K. Metabolic changes in autoimmune diseases. Curr. Drug Discov. Technol. 6, 256–261 (2009).
Bernini, P. et al. Are patients with potential celiac disease really potential? The answer of metabolomics. J. Proteome Res. 10, 714–721 (2011).
El-Hini, S. H. et al. Visfatin and adiponectin as novel markers for evaluation of metabolic disturbance in recently diagnosed rheumatoid arthritis patients. Rheumatol. Int. http://dx.doi.org/10.1007/s00296-013-2714-2713.
Wu, T. et al. Metabolic disturbances associated with systemic lupus erythematosus. PLoS ONE 7, e37210 (2012).
Wang, Z. et al. (1)H NMR-based metabolomic analysis for identifying serum biomarkers to evaluate methotrexate treatment in patients with early rheumatoid arthritis. Exp. Ther. Med. 4, 165–171 (2012).
McCulloch, M. et al. Diagnostic accuracy of canine scent detection in early- and late-stage lung and breast cancers. Integr. Cancer Ther. 5, 30–39 (2006).
Robroeks, C. M. et al. Metabolomics of volatile organic compounds in cystic fibrosis patients and controls. Pediatr. Res. 68, 75–80 (2010).
Van Berkel, J. J. et al. A profile of volatile organic compounds in breath discriminates COPD patients from controls. Respir. Med. 104, 557–563 (2010).
Verdam, F. J. et al. Non-alcoholic steatohepatitis: A non-invasive diagnosis by analysis of exhaled breath. J. Hepatol. 58, 543–548 (2013).
Baranska, A. et al. Profile of volatile organic compounds in exhaled breath changes as a result of gluten-free diet. J. Breath Res. 7, 037104 (2013).
Kamada, N., Seo, S.-U., Chen, G. Y. & Núñez, G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 13, 321–335 (2013).
Zeeuwen, P. L. et al. Microbiome dynamics of human epidermis following skin barrier disruption. Genome Biol. 13, R101 (2012).
Sellitto, M. et al. Proof of concept of microbiome-metabolome analysis and delayed gluten exposure on celiac disease autoimmunity in genetically at-risk infants. PLoS ONE 7, e33387 (2012).
Mori, K., Nakagawa, Y. & Ozaki, H. Does the gut microbiota trigger Hashimoto's thyroiditis? Discov. Med. 14, 321–326 (2012).
Hogen Esch, C. E. et al. The PreventCD Study design: towards new strategies for the prevention of coeliac disease. Eur. J. Gastroenterol. Hepatol. 22, 1424–1430 (2010).
Romanos, J. et al. Improving coeliac disease risk prediction by testing non-HLA variants additional to HLA variants. Gut http://dx.doi.org/10.1136/gutjnl-2012-304110.
Manolio, T. A., Bailey-Wilson, J. E. & Collins, F. S. Genes, environment and the value of prospective cohort studies. Nat. Rev. Genet. 7, 812–820 (2006).
Bluestone, J. A., Herold, K. & Eisenbarth, G. Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature 464, 1293–1300 (2010).
Acknowledgements
The authors would like to acknowledge grants from Top Institute Food and Nutrition, the Netherlands (TiFN GH001 to C. Wijmenga and A. Zhernakova) and from the European Research Council under the European Union's Seventh Framework Programme (FP/2007-2013)/ERC Grant Agreement no. 2012- 322698 to C. Wijmenga, the Dutch Digestive Diseases Foundation (MLDS, WO11-30 to C. Wijmenga), and the Dutch MS Foundation (11-752 MS) to S. Withoff. A. Zhernakova holds a Rosalind Franklin Fellowship from the University of Groningen and a grant from the Dutch Reumafonds (11-1-101).
Author information
Authors and Affiliations
Contributions
All authors contributed equally to all aspects of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figure 1
An overview of the common, less common and rare autoimmune and/or endocrine diseases and their affected organs. (PDF 1305 kb)
Supplementary Table 1
GWAS studies in this Review (XLS 49 kb)
Supplementary Table 2
Ichip studies in this Review (XLS 26 kb)
Supplementary Table 3
Associated loci in disease (XLS 72 kb)
Supplementary Table 4
Selected genes and drug targets (XLS 70 kb)
Rights and permissions
About this article
Cite this article
Zhernakova, A., Withoff, S. & Wijmenga, C. Clinical implications of shared genetics and pathogenesis in autoimmune diseases. Nat Rev Endocrinol 9, 646–659 (2013). https://doi.org/10.1038/nrendo.2013.161
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrendo.2013.161
This article is cited by
-
M6A methylation modification in autoimmune diseases, a promising treatment strategy based on epigenetics
Arthritis Research & Therapy (2023)
-
Research progress of engineered mesenchymal stem cells and their derived exosomes and their application in autoimmune/inflammatory diseases
Stem Cell Research & Therapy (2023)
-
Association of early-onset myasthenia gravis and primary Sjögren’s syndrome: a case-based narrative review
Clinical Rheumatology (2022)
-
Autoimmune encephalitis associated with anti-LGI1 antibody and Sjögren’s syndrome: a case report
Neurological Sciences (2021)
-
Five-year trajectories of multimorbidity patterns in an elderly Mediterranean population using Hidden Markov Models
Scientific Reports (2020)