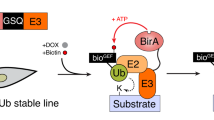
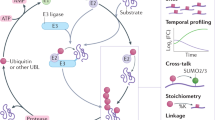
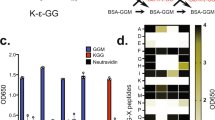
Abstract
Ubiquitin-binding proteins play an important role in eukaryotes by translating differently linked polyubiquitin chains into proper cellular responses. Current knowledge about ubiquitin-binding proteins and ubiquitin linkage-selective interactions is mostly based on case-by-case studies. We have recently reported a method called ubiquitin interactor affinity enrichment–mass spectrometry (UbIA-MS), which enables comprehensive identification of ubiquitin interactors for all ubiquitin linkages from crude cell lysates. One major strength of UbIA-MS is the fact that ubiquitin interactors are enriched from crude cell lysates, in which proteins are present at endogenous levels, contain biologically relevant post-translational modifications (PTMs) and are assembled in native protein complexes. In addition, UbIA-MS uses chemically synthesized nonhydrolyzable diubiquitin, which mimics native diubiquitin and is inert to cleavage by endogenous deubiquitinases (DUBs). Here, we present a detailed protocol for UbIA-MS that proceeds in five stages: (i) chemical synthesis of ubiquitin precursors and click chemistry for the generation of biotinylated nonhydrolyzable diubiquitin baits, (ii) in vitro affinity purification of ubiquitin interactors, (iii) on-bead interactor digestion, (iv) liquid chromatography (LC)–MS/MS analysis and (v) data analysis to identify differentially enriched proteins. The computational analysis tools are freely available as an open-source R software package, including a graphical interface. Typically, UbIA-MS allows the identification of dozens to hundreds of ubiquitin interactors from any type of cell lysate, and can be used to study cell type or stimulus-dependent ubiquitin interactions. The nonhydrolyzable diubiquitin synthesis can be completed in 3 weeks, followed by ubiquitin interactor enrichment and identification, which can be completed within another 2 weeks.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ciechanover, A. The unravelling of the ubiquitin system. Nat. Rev. Mol. Cell Biol. 16, 322–324 (2015).
Husnjak, K. & Dikic, I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 81, 291–322 (2012).
Swatek, K.N. & Komander, D. Ubiquitin modifications. Cell Res. 26, 399–422 (2016).
Dikic, I., Wakatsuki, S. & Walters, K.J. Ubiquitin-binding domains — from structures to functions. Nat. Rev. Mol. Cell Biol. 10, 659–671 (2009).
Zhang, X. et al. An interaction landscape of ubiquitin signaling. Mol. Cell 65, 941–955.e8 (2017).
Rösner, D., Schneider, T., Schneider, D. & Scheffner, M. Click chemistry for targeted protein ubiquitylation and ubiquitin chain formation. Nat. Protoc. 10, 1594–1611 (2015).
Chojnacki, M., Mansour, W., Hameed, D.S. & Singh, R.K. Polyubiquitin-photoactivatable crosslinking reagents for mapping ubiquitin interactome identify Rpn1 as a proteasome ubiquitin-associating subunit. Cell Chem. Biol. 24, 443–457.e6 (2017).
Hurley, J.H., Lee, S. & Prag, G. Ubiquitin-binding domains. Biochem. J. 399, 361–372 (2006).
Geiger, T., Wehner, A., Schaab, C., Cox, J. & Mann, M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteomics 11, M111.014050 (2012).
Kumar, K.S.A., Spasser, L., Erlich, L.A., Bavikar, S.N. & Brik, A. Total chemical synthesis of di-ubiquitin chains. Angew. Chem. Int. Ed. Engl. 49, 9126–9131 (2010).
Weikart, N.D., Sommer, S. & Mootz, H.D. Click synthesis of ubiquitin dimer analogs to interrogate linkage-specific UBA domain binding. Chem. Commun. (Camb.) 48, 296–298 (2012).
El Oualid, F. et al. Chemical synthesis of ubiquitin, ubiquitin-based probes, and diubiquitin. Angew. Chem. Int. Ed. Engl. 49, 10149–10153 (2010).
Eger, S., Scheffner, M., Marx, A. & Rubini, M. Synthesis of defined ubiquitin dimers. J. Am. Chem. Soc. 132, 16337–16339 (2010).
Flierman, D. et al. Non-hydrolyzable diubiquitin probes reveal linkage-specific reactivity of deubiquitylating enzymes mediated by S2 pockets. Cell Chem. Biol. 23, 472–482 (2016).
Kimple, M.E. & Sondek, J. Overview of affinity tags for protein purification. Curr. Protoc. Protein Sci. Chapter 9 Unit 9.9 (2004).
Nielsen, M.L. et al. Iodoacetamide-induced artifact mimics ubiquitination in mass spectrometry. Nat. Methods 5, 459–460 (2008).
Smits, A.H. & Vermeulen, M. Characterizing protein-protein interactions using mass spectrometry: challenges and opportunities. Trends Biotechnol. 34, 825–834 (2016).
Cox, J. et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 (2014).
Cox, J. et al. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 (2009).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016).
Huber, W. et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12, 115–121 (2015).
Huber, W., Von Heydebreck, A. & Sültmann, H. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18, S96–104 (2002).
Gatto, L. & Lilley, K.S. MSnbase-an R/Bioconductor package for isobaric tagged mass spectrometry data visualization, processing and quantitation. Bioinformatics 28, 288–289 (2012).
Smyth, G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, 3 (2004).
Ritchie, M.E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Rappsilber, J., Mann, M. & Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007).
Tammsalu, T. et al. Proteome-wide identification of SUMO modification sites by mass spectrometry. Nat. Protoc. 10, 1374–1388 (2015).
Oualid, F.E., Hameed, D.S. & Atmioui, D.E. Synthesis of atypical diubiquitin chains. Methods Mol. Biol. 832, 597–609 (2012).
Valverde, I.E., Bauman, A. & Kluba, C.A. 1, 2, 3-Triazoles as amide bond mimics: triazole scan yields protease-resistant peptidomimetics for tumor targeting. Angew. Chem. Int. Ed. Engl. 52, 8957–8960 (2013).
Zhou, Z. & Fahrni, C.J. A fluorogenic probe for the copper(I)-catalyzed azide-alkyne ligation reaction: modulation of the fluorescence emission via 3(n,pi)-1(pi,pi) inversion. J. Am. Chem. Soc. 126, 8862–8863 (2004).
Acknowledgements
We thank B. Klaus for statistical advice and members of the Vermeulen and Ovaa labs for fruitful discussions. Work in the Zhang lab was supported by the “Hundred Talents Program C” of the Chinese Academy of Sciences (no. 2017-045) and by Guangdong Science and Technology Projects (2014B050504008, 2014B050502012, 2014B020225002 and 2014B030301058). Work in the Vermeulen lab was supported by the NWO Gravitation program CGC.nl. Work in the Ovaa lab was supported by the ERC grant Ubicode (no. 281699). A.H.S. was supported by a fellowship from the EMBL Interdisciplinary Postdoc (EIPOD) Programme under a grant from the Marie Sklodowska-Curie Actions COFUND (no. 664726).
Author information
Authors and Affiliations
Contributions
X.Z. and M.V. designed the method with input from A.H.S., G.B.A.v.T., W.H. and H.O. A.H.S. prepared figures, and developed and maintained the R package. X.Z., A.H.S., G.B.A.v.T. and M.V. wrote the manuscript with input from H.O. and W.H.
Corresponding authors
Ethics declarations
Competing interests
H.O. is a shareholder in the biotechnology company UbiQ. The other authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Quality-control plots.
(a) The number of proteins identified in every sample. Colors indicate the conditions and the solid line indicates the number of proteins identified in all samples (b) Stacked barplot to visualize the total number of proteins classified by the number of samples they are identified in (stacks and colors). (c) Barplot to visualize the number of proteins identified classified by the number of samples they are identified in (colors). (d) Boxplots of protein intensity distributions before (bottom) and after (top) normalization. (e) Heatmap of the proteins with missing values. Colors indicate whether the values are missing (white) or are valid (black).
Supplementary Figure 2 Heatmap of linkage-selective interactors of diubiquitin linkages in HeLa cells.
Layout, clustering and colors are as in Figure 3.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2. (PDF 795 kb)
Rights and permissions
About this article
Cite this article
Zhang, X., Smits, A., van Tilburg, G. et al. Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat Protoc 13, 530–550 (2018). https://doi.org/10.1038/nprot.2017.147
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2017.147
This article is cited by
-
Targeting cancer-derived extracellular vesicles by combining CD147 inhibition with tissue factor pathway inhibitor for the management of urothelial cancer cells
Cell Communication and Signaling (2024)
-
The TP53-activated E3 ligase RNF144B is a tumour suppressor that prevents genomic instability
Journal of Experimental & Clinical Cancer Research (2024)
-
FOXO1-mediated lipid metabolism maintains mammalian embryos in dormancy
Nature Cell Biology (2024)
-
Differentiation granules, a dynamic regulator of T. brucei development
Nature Communications (2024)
-
Rewiring of the phosphoproteome executes two meiotic divisions in budding yeast
The EMBO Journal (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.