Abstract
The modification of proteins by the addition of ubiquitin and other ubiquitin-like proteins (UBLs) is involved in a wide range of cellular processes including cell cycle progression, the DNA damage response, endocytosis, cell signalling, autophagy and protein quality control. The UBL family comprises more than a dozen structurally related members, with ubiquitin, small ubiquitin-like modifier (SUMO) proteins, NEDD8, ISG15 and FAT10 being the most commonly known. Each UBL is associated with a distinct set of enzymes that alter the architecture and fate of their cognate proteins. UBL-conjugating enzymes add one or more UBLs to lysine and non-lysine acceptor sites on their target proteins, forming a complex distribution of monomeric and polymeric modifications. Different approaches and strategies are available to identify the sites of UBL modification, the types of modification and their dynamics upon various cellular stimuli; these techniques can decipher the complex architecture of UBL substrates and expand our understanding of UBL functions and their importance in cellular homeostasis and human diseases. This Primer covers the current methods for identifying UBL substrates, their modification sites and UBL chain linkages, and describes where the application of these methods can be used to gain biological insights into UBL functions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 1 digital issues and online access to articles
$99.00 per year
only $99.00 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Rajalingam, K. & Dikic, I. SnapShot: expanding the ubiquitin code. Cell 164, 1074–1074.e1 (2016).
Yau, R. & Rape, M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 18, 579–586 (2016). This review highlights the diversity of ubiquitin chain topologies and their impact on cell signalling.
Kerscher, O., Felberbaum, R. & Hochstrasser, M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180 (2006).
Cappadocia, L. & Lima, C. D. Ubiquitin-like protein conjugation: structures, chemistry, and mechanism. Chem. Rev. 118, 889–918 (2018). This Review describes the conjugation machinery of UBLs and different approaches to identify intermediates during UBL activation and conjugation.
Clague, M. J., Urbe, S. & Komander, D. Breaking the chains: deubiquitylating enzyme specificity begets function. Nat. Rev. Mol. Cell Biol. 20, 338–352 (2019).
Ronau, J. A., Beckmann, J. F. & Hochstrasser, M. Substrate specificity of the ubiquitin and Ubl proteases. Cell Res. 26, 441–456 (2016).
McClellan, A. J., Laugesen, S. H. & Ellgaard, L. Cellular functions and molecular mechanisms of non-lysine ubiquitination. Open. Biol. 9, 190147 (2019).
Jansen, N. S. & Vertegaal, A. C. O. A chain of events: regulating target proteins by SUMO polymers. Trends Biochem. Sci. 46, 113–123 (2021).
Bailly, A. P. et al. The balance between mono- and NEDD8-chains controlled by NEDP1 upon DNA damage is a regulatory module of the HSP70 ATPase activity. Cell Rep. 29, 212–224.e8 (2019).
Keuss, M. J. et al. Unanchored tri-NEDD8 inhibits PARP-1 to protect from oxidative stress-induced cell death. EMBO J. 38, e100024 (2019).
Perez Berrocal, D. A., Witting, K. F., Ovaa, H. & Mulder, M. P. C. Hybrid chains: a collaboration of ubiquitin and ubiquitin-like modifiers introducing cross-functionality to the ubiquitin code. Front. Chem. 7, 931 (2019).
Streich, F. C. Jr. & Lima, C. D. Structural and functional insights to ubiquitin-like protein conjugation. Annu. Rev. Biophys. 43, 357–379 (2014).
van der Veen, A. G. & Ploegh, H. L. Ubiquitin-like proteins. Annu. Rev. Biochem. 81, 323–357 (2012).
Akutsu, M., Dikic, I. & Bremm, A. Ubiquitin chain diversity at a glance. J. Cell Sci. 129, 875–880 (2016). This paper presents a method to unravel the complex architecture of ubiquitin chains to understand polyubiquitin signals.
Swatek, K. N. et al. Insights into ubiquitin chain architecture using Ub-clipping. Nature 572, 533–537 (2019).
El-Asmi, F. et al. Cross-talk between SUMOylation and ISGylation in response to interferon. Cytokine 129, 155025 (2020).
Aichem, A. et al. The proteomic analysis of endogenous FAT10 substrates identifies p62/SQSTM1 as a substrate of FAT10ylation. J. Cell Sci. 125, 4576–4585 (2012).
McManus, F. P., Lamoliatte, F. & Thibault, P. Identification of cross talk between SUMOylation and ubiquitylation using a sequential peptide immunopurification approach. Nat. Protoc. 12, 2342–2358 (2017).
Udeshi, N. D., Mertins, P., Svinkina, T. & Carr, S. A. Large-scale identification of ubiquitination sites by mass spectrometry. Nat. Protoc. 8, 1950–1960 (2013).
Hendriks, I. A. & Vertegaal, A. C. A high-yield double-purification proteomics strategy for the identification of SUMO sites. Nat. Protoc. 11, 1630–1649 (2016).
Barysch, S. V., Dittner, C., Flotho, A., Becker, J. & Melchior, F. Identification and analysis of endogenous SUMO1 and SUMO2/3 targets in mammalian cells and tissues using monoclonal antibodies. Nat. Protoc. 9, 896–909 (2014).
Liu, N. et al. Clinically used antirheumatic agent auranofin is a proteasomal deubiquitinase inhibitor and inhibits tumor growth. Oncotarget 5, 5453–5471 (2014).
Liu, N. et al. A novel proteasome inhibitor suppresses tumor growth via targeting both 19S proteasome deubiquitinases and 20S proteolytic peptidases. Sci. Rep. 4, 5240 (2014).
Zhang, J. J., Ng, K. M., Lok, C. N., Sun, R. W. & Che, C. M. Deubiquitinases as potential anti-cancer targets for gold(III) complexes. Chem. Commun. 49, 5153–5155 (2013).
Graham, J. M. Fractionation of subcellular organelles. Curr. Protoc. Cell Biol. 69, 3.1.1–3.1.22 (2015).
Wagner, S. A. et al. Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol. Cell Proteom. 11, 1578–1585 (2012).
McManus, F. P., Altamirano, C. D. & Thibault, P. In vitro assay to determine SUMOylation sites on protein substrates. Nat. Protoc. 11, 387–397 (2016).
Jeram, S. M., Srikumar, T., Pedrioli, P. G. & Raught, B. Using mass spectrometry to identify ubiquitin and ubiquitin-like protein conjugation sites. Proteomics 9, 922–934 (2009).
Argenzio, E. et al. Proteomic snapshot of the EGF-induced ubiquitin network. Mol. Syst. Biol. 7, 462 (2011).
Danielsen, J. M. et al. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol. Cell Proteom. 10, M110 003590 (2011).
Peng, J. et al. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 (2003).
Roux, K. J., Kim, D. I., Burke, B. & May, D. G. BioID: a screen for protein–protein interactions. Curr. Protoc. Protein Sci. 91, 19.23.11–19.23.15 (2018).
Hill, Z. B., Pollock, S. B., Zhuang, M. & Wells, J. A. Direct proximity tagging of small molecule protein targets using an engineered NEDD8 ligase. J. Am. Chem. Soc. 138, 13123–13126 (2016).
Coyaud, E. et al. BioID-based identification of Skp Cullin F-box (SCF)β–TrCP1/2 E3 ligase substrates. Mol. Cell Proteom. 14, 1781–1795 (2015).
Branon, T. C. et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887 (2018).
Hendriks, I. A. et al. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 21, 927–936 (2014). This paper describes a method to purify SUMOylated proteins, enabling the identification of more than 4,300 SUMOylation sites in human cells.
Impens, F., Radoshevich, L., Cossart, P. & Ribet, D. Mapping of SUMO sites and analysis of SUMOylation changes induced by external stimuli. Proc. Natl Acad. Sci. USA 111, 12432–12437 (2014).
Lamoliatte, F. et al. Large-scale analysis of lysine SUMOylation by SUMO remnant immunoaffinity profiling. Nat. Commun. 5, 5409 (2014).
Tammsalu, T. et al. Proteome-wide identification of SUMO2 modification sites. Sci. Signal. 7, rs2 (2014).
O’Connor, H. F. et al. Ubiquitin-activated interaction traps (UBAITs) identify E3 ligase binding partners. EMBO Rep. 16, 1699–1712 (2015).
Kumar, R., Gonzalez-Prieto, R., Xiao, Z., Verlaan-de Vries, M. & Vertegaal, A. C. O. The STUbL RNF4 regulates protein group SUMOylation by targeting the SUMO conjugation machinery. Nat. Commun. 8, 1809 (2017).
Hjerpe, R. et al. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 10, 1250–1258 (2009).
Yoshida, Y. et al. A comprehensive method for detecting ubiquitinated substrates using TR-TUBE. Proc. Natl Acad. Sci. USA 112, 4630–4635 (2015).
Gao, Y. et al. Enhanced purification of ubiquitinated proteins by engineered tandem hybrid ubiquitin-binding domains (ThUBDs). Mol. Cell Proteom. 15, 1381–1396 (2016).
Lang, V., Da Silva-Ferrada, E., Barrio, R., Sutherland, J. D. & Rodriguez, M. S. Using biotinylated SUMO-traps to analyze SUMOylated proteins. Methods Mol. Biol. 1475, 109–121 (2016).
Da Silva-Ferrada, E. et al. Analysis of SUMOylated proteins using SUMO-traps. Sci. Rep. 3, 1690 (2013).
Zhuang, M., Guan, S., Wang, H., Burlingame, A. L. & Wells, J. A. Substrates of IAP ubiquitin ligases identified with a designed orthogonal E3 ligase, the NEDDylator. Mol. Cell 49, 273–282 (2013).
Salas-Lloret, D., Agabitini, G. & Gonzalez-Prieto, R. TULIP2: an improved method for the identification of ubiquitin E3-specific targets. Front. Chem. 7, 802 (2019).
Khoshnood, B., Dacklin, I. & Grabbe, C. A proteomics approach to identify targets of the ubiquitin-like molecule Urm1 in Drosophila melanogaster. PLoS ONE 12, e0185611 (2017).
Leng, L. et al. A proteomics strategy for the identification of FAT10-modified sites by mass spectrometry. J. Proteome Res. 13, 268–276 (2014).
Yoo, H. M. et al. Modification of ASC1 by UFM1 is crucial for ERα transactivation and breast cancer development. Mol. Cell 56, 261–274 (2014).
Bakos, G. et al. An E2-ubiquitin thioester-driven approach to identify substrates modified with ubiquitin and ubiquitin-like molecules. Nat. Commun. 9, 4776 (2018).
Uzoma, I. et al. Global identification of small ubiquitin-related modifier (SUMO) substrates reveals crosstalk between SUMOylation and phosphorylation promotes cell migration. Mol. Cell Proteom. 17, 871–888 (2018).
Merbl, Y., Refour, P., Patel, H., Springer, M. & Kirschner, M. W. Profiling of ubiquitin-like modifications reveals features of mitotic control. Cell 152, 1160–1172 (2013).
Oh, Y. H. et al. Chip-based analysis of SUMO (small ubiquitin-like modifier) conjugation to a target protein. Biosens. Bioelectron. 22, 1260–1267 (2007).
Akimov, V. et al. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 25, 631–640 (2018). This paper presents the UbiSite immunoaffinity method to specifically enrich and identify ubiquitin-modified proteins uncovering more than 63,000 ubiquitylation sites on 9,200 proteins.
Xu, P. & Peng, J. Characterization of polyubiquitin chain structure by middle-down mass spectrometry. Anal. Chem. 80, 3438–3444 (2008).
Vogl, A. M. et al. Global site-specific neddylation profiling reveals that NEDDylated cofilin regulates actin dynamics. Nat. Struct. Mol. Biol. 27, 210–220 (2020).
Hendriks, I. A. et al. Site-specific characterization of endogenous SUMOylation across species and organs. Nat. Commun. 9, 2456 (2018).
Rinfret Robert, C., McManus, F. P., Lamoliatte, F. & Thibault, P. Interplay of ubiquitin-like modifiers following arsenic trioxide treatment. J. Proteome Res. 19, 1999–2010 (2020).
Lamoliatte, F., McManus, F. P., Maarifi, G., Chelbi-Alix, M. K. & Thibault, P. Uncovering the SUMOylation and ubiquitylation crosstalk in human cells using sequential peptide immunopurification. Nat. Commun. 8, 14109 (2017). This paper uses a dual-remnant immunoaffinity enrichment to identify protein SUMOylation and ubiquitylation in a site-specific manner.
Fulzele, A. & Bennett, E. J. Ubiquitin diGLY proteomics as an approach to identify and quantify the ubiquitin-modified proteome. Methods Mol. Biol. 1844, 363–384 (2018).
Lumpkin, R. J. et al. Site-specific identification and quantitation of endogenous SUMO modifications under native conditions. Nat. Commun. 8, 1171 (2017). This paper features a method to enrich endogenous SUMOylated proteins and their identification by mass spectrometry, revealing more than 1,200 SUMO sites.
Cai, L. et al. Proteome-wide mapping of endogenous SUMOylation sites in mouse testis. Mol. Cell Proteom. 16, 717–727 (2017).
Ludwig, C. et al. Data-independent acquisition-based SWATH-MS for quantitative proteomics: a tutorial. Mol. Syst. Biol. 14, e8126 (2018).
Hansen, F. M. et al. Data-independent acquisition method for ubiquitinome analysis reveals regulation of circadian biology. Nat. Commun. 12, 254 (2021).
Calderon-Celis, F., Encinar, J. R. & Sanz-Medel, A. Standardization approaches in absolute quantitative proteomics with mass spectrometry. Mass. Spectrom. Rev. 37, 715–737 (2018).
Cox, J. & Mann, M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 80, 273–299 (2011).
Ong, S. E. & Mann, M. Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 (2005).
Rodriguez-Suarez, E. & Whetton, A. D. The application of quantification techniques in proteomics for biomedical research. Mass. Spectrom. Rev. 32, 1–26 (2013).
Cox, J. et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteom. 13, 2513–2526 (2014).
Lear, T. B. et al. Kelch-like protein 42 is a profibrotic ubiquitin E3 ligase involved in systemic sclerosis. J. Biol. Chem. 295, 4171–4180 (2020).
Zhu, W., Smith, J. W. & Huang, C. M. Mass spectrometry-based label-free quantitative proteomics. J. Biomed. Biotechnol. 2010, 840518 (2010).
Hogrebe, A. et al. Benchmarking common quantification strategies for large-scale phosphoproteomics. Nat. Commun. 9, 1045 (2018).
Ong, S. E. et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteom. 1, 376–386 (2002).
An, J. et al. pSILAC mass spectrometry reveals ZFP91 as IMiD-dependent substrate of the CRL4(CRBN) ubiquitin ligase. Nat. Commun. 8, 15398 (2017).
Geiger, T., Cox, J., Ostasiewicz, P., Wisniewski, J. R. & Mann, M. Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat. Methods 7, 383–385 (2010).
Wiese, S., Reidegeld, K. A., Meyer, H. E. & Warscheid, B. Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics 7, 340–350 (2007).
Thompson, A. et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 (2003).
Rose, C. M. et al. Highly multiplexed quantitative mass spectrometry analysis of ubiquitylomes. Cell Syst. 3, 395–403.e4 (2016).
Niu, M. et al. Extensive peptide fractionation and y1 ion-based interference detection method for enabling accurate quantification by isobaric labeling and mass spectrometry. Anal. Chem. 89, 2956–2963 (2017).
Wuhr, M. et al. Accurate multiplexed proteomics at the MS2 level using the complement reporter ion cluster. Anal. Chem. 84, 9214–9221 (2012).
Ting, L., Rad, R., Gygi, S. P. & Haas, W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods 8, 937–940 (2011).
Udeshi, N. D. et al. Rapid and deep-scale ubiquitylation profiling for biology and translational research. Nat. Commun. 11, 359 (2020).
Perkins, D. N., Pappin, D. J., Creasy, D. M. & Cottrell, J. S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999).
Eng, J. K., McCormack, A. L. & Yates, J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass. Spectrom. 5, 976–989 (1994).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Zhang, J. et al. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell Proteom. 11, M111 010587 (2012).
Li, Y. et al. An integrated bioinformatics platform for investigating the human E3 ubiquitin ligase–substrate interaction network. Nat. Commun. 8, 347 (2017).
Xue, Y., Zhou, F., Fu, C., Xu, Y. & Yao, X. SUMOsp: a web server for sumoylation site prediction. Nucleic Acids Res. 34, W254–W257 (2006).
Ren, J. et al. Systematic study of protein sumoylation: development of a site-specific predictor of SUMOsp 2.0. Proteomics 9, 3409–3412 (2009).
Xue, Y. et al. GPS: a comprehensive www server for phosphorylation sites prediction. Nucleic Acids Res. 33, W184–W187 (2005).
Schwartz, D. & Gygi, S. P. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 (2005).
Zhao, Q. et al. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 42, W325–W330 (2014).
Beauclair, G., Bridier-Nahmias, A., Zagury, J. F., Saib, A. & Zamborlini, A. JASSA: a comprehensive tool for prediction of SUMOylation sites and SIMs. Bioinformatics 31, 3483–3491 (2015).
Dehzangi, A., Lopez, Y., Taherzadeh, G., Sharma, A. & Tsunoda, T. SumSec: accurate prediction of sumoylation sites using predicted secondary structure. Molecules 23, 3260 (2018).
Sharma, A. et al. HseSUMO: sumoylation site prediction using half-sphere exposures of amino acids residues. BMC Genomics 19, 982 (2019).
Qian, Y., Ye, S., Zhang, Y. & Zhang, J. SUMO-Forest: a cascade forest based method for the prediction of SUMOylation sites on imbalanced data. Gene 741, 144536 (2020).
Xu, H. D., Liang, R. P., Wang, Y. G. & Qiu, J. D. mUSP: a high-accuracy map of the in situ crosstalk of ubiquitylation and SUMOylation proteome predicted via the feature enhancement approach. Brief. Bioinform. 22, bbaa050 (2021).
Qiu, W., Xu, C., Xiao, X. & Xu, D. Computational prediction of ubiquitination proteins using evolutionary profiles and functional domain annotation. Curr. Genomics 20, 389–399 (2019).
He, F. et al. Large-scale prediction of protein ubiquitination sites using a multimodal deep architecture. BMC Syst. Biol. 12, 109 (2018).
Yavuz, A. S., Sozer, N. B. & Sezerman, O. U. Prediction of neddylation sites from protein sequences and sequence-derived properties. BMC Bioinformatics 16 (Suppl. 18), S9 (2015).
Hornbeck, P. V. et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 (2012).
Tammsalu, T. et al. Proteome-wide identification of SUMO modification sites by mass spectrometry. Nat. Protoc. 10, 1374–1388 (2015).
Cox, J. et al. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat.Protoc. 4, 698–705 (2009).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016).
Ma, B. et al. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass. Spectrom. 17, 2337–2342 (2003).
Hendriks, I. A. et al. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat. Struct. Mol. Biol. 24, 325–336 (2017).
Chachami, G. et al. Hypoxia-induced changes in SUMO conjugation affect transcriptional regulation under low oxygen. Mol. Cell Proteom. 18, 1197–1209 (2019).
Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016).
Fu, H., Yang, Y., Wang, X., Wang, H. & Xu, Y. DeepUbi: a deep learning framework for prediction of ubiquitination sites in proteins. BMC Bioinformatics 20, 86 (2019).
Liu, Y., Li, A., Zhao, X. M. & Wang, M. DeepTL-Ubi: a novel deep transfer learning method for effectively predicting ubiquitination sites of multiple species. Methods 192, 103–111 (2020).
The Gene Ontology Consortium. The Gene Ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338 (2019).
Eifler, K. et al. SUMO targets the APC/C to regulate transition from metaphase to anaphase. Nat. Commun. 9, 1119 (2018).
El-Gebali, S. et al. The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432 (2019).
Szklarczyk, D. et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Psakhye, I. & Jentsch, S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151, 807–820 (2012). This paper highlights the concept of the simultaneous modification of related sets of target proteins to regulate cellular processes by PTM.
Schwertman, P., Bekker-Jensen, S. & Mailand, N. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat. Rev. Mol. Cell Biol. 17, 379–394 (2016).
Jackson, S. P. & Durocher, D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol.Cell 49, 795–807 (2013).
Odeh, H. M., Coyaud, E., Raught, B. & Matunis, M. J. The SUMO-specific isopeptidase SENP2 is targeted to intracellular membranes via a predicted N-terminal amphipathic α-helix. Mol. Biol. Cell 29, 1878–1890 (2018).
Xu, P. et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145 (2009).
Kim, W. et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 (2011).
Xu, G., Paige, J. S. & Jaffrey, S. R. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol. 28, 868–873 (2010). This study is the first to report on the use of diglycine remnant immunopurification for large-scale identification of ubiquitin-modified proteomes.
Zhang, Y. et al. The in vivo ISGylome links ISG15 to metabolic pathways and autophagy upon Listeria monocytogenes infection. Nat. Commun. 10, 5383 (2019).
Pinto-Fernandez, A. et al. Deletion of the deISGylating enzyme USP18 enhances tumour cell antigenicity and radiosensitivity. Br. J. Cancer 124, 817–830 (2021).
Steen, H. & Mann, M. The ABC’s (and XYZ’s) of peptide sequencing. Nat. Rev. Mol. Cell Biol. 5, 699–711 (2004).
Na, C. H. et al. Synaptic protein ubiquitination in rat brain revealed by antibody-based ubiquitome analysis. J. Proteome Res. 11, 4722–4732 (2012).
Griffin, N. M. et al. Label-free, normalized quantification of complex mass spectrometry data for proteomic analysis. Nat. Biotechnol. 28, 83–89 (2010).
Murie, C. et al. Normalization of mass spectrometry data (NOMAD). Adv. Biol. Regul. 67, 128–133 (2018).
Vertegaal, A. C. Uncovering ubiquitin and ubiquitin-like signaling networks. Chem. Rev. 111, 7923–7940 (2011).
Aichem, A. et al. The ubiquitin-like modifier FAT10 interferes with SUMO activation. Nat. Commun. 10, 4452 (2019).
French, M. E., Koehler, C. F. & Hunter, T. Emerging functions of branched ubiquitin chains. Cell Discov. 7, 6 (2021).
Haakonsen, D. L. & Rape, M. Branching out: improved signaling by heterotypic ubiquitin chains. Trends Cell Biol. 29, 704–716 (2019).
Kane, L. A. et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 205, 143–153 (2014).
Kazlauskaite, A. et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 460, 127–139 (2014).
Koyano, F. et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166 (2014). In this study, phosphorylation of ubiquitin at Ser65 by PINK1 is found to enhance ubiquitin conjugation by the E3 ligase Parkin.
Ordureau, A. et al. Dynamics of PARKIN-dependent mitochondrial ubiquitylation in induced neurons and model systems revealed by digital snapshot proteomics. Mol. Cell 70, 211–227.e8 (2018).
Wauer, T. et al. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 34, 307–325 (2015).
Baek, K. et al. NEDD8 nucleates a multivalent Cullin–RING–UBE2D ubiquitin ligation assembly. Nature 578, 461–466 (2020).
Da Costa, I. C. & Schmidt, C. K. Ubiquitin-like proteins in the DNA damage response: the next generation. Essays Biochem. 64, 737–752 (2020).
Uzunova, K. et al. Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 282, 34167–34175 (2007).
Cuijpers, S. A. G., Willemstein, E. & Vertegaal, A. C. O. Converging small ubiquitin-like modifier (SUMO) and ubiquitin signaling: improved methodology identifies co-modified target proteins. Mol. Cell Proteom. 16, 2281–2295 (2017).
Pichler, A. et al. SUMO modification of the ubiquitin-conjugating enzyme E2-25K. Nat. Struct. Mol. Biol. 12, 264–269 (2005).
Ranieri, M. et al. Sumoylation and ubiquitylation crosstalk in the control of ΔNp63α protein stability. Gene 645, 34–40 (2018).
Lecona, E. et al. USP7 is a SUMO deubiquitinase essential for DNA replication. Nat. Struct. Mol. Biol. 23, 270–277 (2016).
Hendriks, I. A. & Vertegaal, A. C. A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 17, 581–595 (2016).
Prudden, J. et al. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 26, 4089–4101 (2007).
Sun, H., Leverson, J. D. & Hunter, T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 26, 4102–4112 (2007).
Poulsen, S. L. et al. RNF111/Arkadia is a SUMO-targeted ubiquitin ligase that facilitates the DNA damage response. J. Cell Biol. 201, 797–807 (2013).
Seenivasan, R. et al. Mechanism and chain specificity of RNF216/TRIAD3, the ubiquitin ligase mutated in Gordon Holmes syndrome. Hum. Mol. Genet. 28, 2862–2873 (2019).
Sriramachandran, A. M. et al. Arkadia/RNF111 is a SUMO-targeted ubiquitin ligase with preference for substrates marked with SUMO1-capped SUMO2/3 chain. Nat. Commun. 10, 3678 (2019).
Yin, Y. et al. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 26, 1196–1208 (2012).
van Cuijk, L. et al. SUMO and ubiquitin-dependent XPC exchange drives nucleotide excision repair. Nat. Commun. 6, 7499 (2015).
Enchev, R. I., Schulman, B. A. & Peter, M. Protein neddylation: beyond Cullin–RING ligases. Nat. Rev. Mol. Cell Biol. 16, 30–44 (2015).
Hicke, L., Schubert, H. L. & Hill, C. P. Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 6, 610–621 (2005).
Husnjak, K. & Dikic, I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 81, 291–322 (2012).
Aksnes, H., Ree, R. & Arnesen, T. Co-translational, post-translational, and non-catalytic roles of N-terminal acetyltransferases. Mol. Cell 73, 1097–1114 (2019).
Bloom, J., Amador, V., Bartolini, F., DeMartino, G. & Pagano, M. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell 115, 71–82 (2003).
Choe, K. N. & Moldovan, G. L. Forging ahead through darkness: PCNA, still the principal conductor at the replication fork. Mol. Cell 65, 380–392 (2017).
Weisshaar, S. R. et al. Arsenic trioxide stimulates SUMO-2/3 modification leading to RNF4-dependent proteolytic targeting of PML. FEBS Lett. 582, 3174–3178 (2008).
Lee, H. S., Lim, Y. S., Park, E. M., Baek, S. H. & Hwang, S. B. SUMOylation of nonstructural 5A protein regulates hepatitis C virus replication. J. Viral Hepat. 21, e108–e117 (2014).
McManus, F. P. et al. Quantitative SUMO proteomics reveals the modulation of several PML nuclear body associated proteins and an anti-senescence function of UBC9. Sci. Rep. 8, 7754 (2018).
Takeuchi, T., Iwahara, S., Saeki, Y., Sasajima, H. & Yokosawa, H. Link between the ubiquitin conjugation system and the ISG15 conjugation system: ISG15 conjugation to the UbcH6 ubiquitin E2 enzyme. J. Biochem. 138, 711–719 (2005).
Takeuchi, T. & Yokosawa, H. ISG15 modification of Ubc13 suppresses its ubiquitin-conjugating activity. Biochem. Biophys. Res. Commun. 336, 9–13 (2005).
Bialas, J., Groettrup, M. & Aichem, A. Conjugation of the ubiquitin activating enzyme UBE1 with the ubiquitin-like modifier FAT10 targets it for proteasomal degradation. PLoS ONE 10, e0120329 (2015).
Mukhopadhyay, D. & Riezman, H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315, 201–205 (2007).
Kliza, K. et al. Internally tagged ubiquitin: a tool to identify linear polyubiquitin-modified proteins by mass spectrometry. Nat. Methods 14, 504–512 (2017).
Michel, M. A., Swatek, K. N., Hospenthal, M. K. & Komander, D. Ubiquitin linkage-specific affimers reveal insights into K6-linked ubiquitin signaling. Mol. Cell 68, 233–246.e5 (2017).
Ordureau, A. et al. Global landscape and dynamics of Parkin and USP30-dependent ubiquitylomes in ineurons during mitophagic signaling. Mol. Cell 77, 1124–1142.e10 (2020).
Yau, R. G. et al. Assembly and function of heterotypic ubiquitin chains in cell-cycle and protein quality control. Cell 171, 918–933 e920 (2017). This paper describes how complex mixed types of ubiquitin polymers play a role in cell cycle progression and protein quality control.
Hendriks, I. A., Schimmel, J., Eifler, K., Olsen, J. V. & Vertegaal, A. C. Ubiquitin-specific protease 11 (USP11) deubiquitinates hybrid small ubiquitin-like modifier (SUMO)-ubiquitin chains to counteract RING finger protein 4 (RNF4). J. Biol. Chem. 290, 15526–15537 (2015).
Guzzo, C. M. et al. RNF4-dependent hybrid SUMO-ubiquitin chains are signals for RAP80 and thereby mediate the recruitment of BRCA1 to sites of DNA damage. Sci. Signal. 5, ra88 (2012).
Maghames, C. M. et al. NEDDylation promotes nuclear protein aggregation and protects the ubiquitin proteasome system upon proteotoxic stress. Nat. Commun. 9, 4376 (2018).
Leidecker, O., Matic, I., Mahata, B., Pion, E. & Xirodimas, D. P. The ubiquitin E1 enzyme Ube1 mediates NEDD8 activation under diverse stress conditions. Cell Cycle 11, 1142–1150 (2012).
Skaug, B. & Chen, Z. J. Emerging role of ISG15 in antiviral immunity. Cell 143, 187–190 (2010).
Fan, J. B. et al. Identification and characterization of a novel ISG15–ubiquitin mixed chain and its role in regulating protein homeostasis. Sci. Rep. 5, 12704 (2015).
Zhao, C., Denison, C., Huibregtse, J. M., Gygi, S. & Krug, R. M. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc. Natl Acad. Sci. USA 102, 10200–10205 (2005).
Anania, V. G. et al. Peptide level immunoaffinity enrichment enhances ubiquitination site identification on individual proteins. Mol. Cell Proteom. 13, 145–156 (2014).
Phu, L. et al. Dynamic regulation of mitochondrial import by the ubiquitin system. Mol. Cell 77, 1107–1123.e10 (2020).
Huang, E. Y. et al. A VCP inhibitor substrate trapping approach (VISTA) enables proteomic profiling of endogenous ERAD substrates. Mol. Biol. Cell 29, 1021–1030 (2018).
Perez-Riverol, Y. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019).
Wang, M. et al. Assembling the community-scale discoverable human proteome. Cell Syst. 7, 412–421.e5 (2018).
Okuda, S. et al. jPOSTrepo: an international standard data repository for proteomes. Nucleic Acids Res. 45, D1107–D1111 (2017).
Ma, J. et al. iProX: an integrated proteome resource. Nucleic Acids Res. 47, D1211–D1217 (2019).
Deutsch, E. W., Lam, H. & Aebersold, R. PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 9, 429–434 (2008).
Sharma, V. et al. Panorama public: a public repository for quantitative data sets processed in skyline. Mol. Cell Proteom. 17, 1239–1244 (2018).
Jentsch, S. & Psakhye, I. Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu. Rev. Genet. 47, 167–186 (2013).
Hewings, D. S., Flygare, J. A., Bogyo, M. & Wertz, I. E. Activity-based probes for the ubiquitin conjugation–deconjugation machinery: new chemistries, new tools, and new insights. FEBS J. 284, 1555–1576 (2017).
Ovaa, H. & Vertegaal, A. C. O. Probing ubiquitin and SUMO conjugation and deconjugation. Biochem. Soc. Trans. 46, 423–436 (2018).
Gui, W. et al. Cell-permeable activity-based ubiquitin probes enable intracellular profiling of human deubiquitinases. J. Am. Chem. Soc. 140, 12424–12433 (2018).
Swatek, K. N. & Komander, D. Ubiquitin modifications. Cell Res. 26, 399–422 (2016).
Gomes, F. et al. Top-down analysis of novel synthetic branched proteins. J. Mass. Spectrom. 54, 19–25 (2019).
Olsen, J. V. & Mann, M. Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol. Cell Proteom. 12, 3444–3452 (2013).
Nielsen, M. L. et al. Iodoacetamide-induced artifact mimics ubiquitination in mass spectrometry. Nat. Methods 5, 459–460 (2008).
Udeshi, N. D. et al. Methods for quantification of in vivo changes in protein ubiquitination following proteasome and deubiquitinase inhibition. Mol. Cell Proteom. 11, 148–159 (2012).
Humphrey, S. J., Karayel, O., James, D. E. & Mann, M. High-throughput and high-sensitivity phosphoproteomics with the EasyPhos platform. Nat. Protoc. 13, 1897–1916 (2018).
Tran, J. C. et al. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature 480, 254–258 (2011).
Lee, A. E. et al. Preparing to read the ubiquitin code: top-down analysis of unanchored ubiquitin tetramers. J. Mass. Spectrom. 51, 629–637 (2016).
Mattern, M., Sutherland, J., Kadimisetty, K., Barrio, R. & Rodriguez, M. S. Using ubiquitin binders to decipher the ubiquitin code. Trends Biochem. Sci. 44, 599–615 (2019).
Huang, X. & Dixit, V. M. Drugging the undruggables: exploring the ubiquitin system for drug development. Cell Res. 26, 484–498 (2016).
Verma, R., Mohl, D. & Deshaies, R. J. Harnessing the power of proteolysis for targeted protein inactivation. Mol. Cell 77, 446–460 (2020).
Karayel, O., Michaelis, A. C., Mann, M., Schulman, B. A. & Langlois, C. R. DIA-based systems biology approach unveils E3 ubiquitin ligase-dependent responses to a metabolic shift. Proc. Natl Acad. Sci. USA 117, 32806–32815 (2020).
Mark, K. G., Loveless, T. B. & Toczyski, D. P. Isolation of ubiquitinated substrates by tandem affinity purification of E3 ligase-polyubiquitin-binding domain fusions (ligase traps). Nat. Protoc. 11, 291–301 (2016).
Kliza, K. & Husnjak, K. Resolving the complexity of ubiquitin networks. Front. Mol. Biosci. 7, 21 (2020).
Li, W. et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS ONE 3, e1487 (2008).
O’Connor, H. F. & Huibregtse, J. M. Enzyme–substrate relationships in the ubiquitin system: approaches for identifying substrates of ubiquitin ligases. Cell Mol. Life Sci. 74, 3363–3375 (2017).
Liebelt, F. et al. The poly-SUMO2/3 protease SENP6 enables assembly of the constitutive centromere-associated network by group deSUMOylation. Nat. Commun. 10, 3987 (2019).
Andaluz Aguilar, H., Iliuk, A. B., Chen, I. H. & Tao, W. A. Sequential phosphoproteomics and N-glycoproteomics of plasma-derived extracellular vesicles. Nat. Protoc. 15, 161–180 (2020).
Melo-Braga, M. N., Ibanez-Vea, M., Larsen, M. R. & Kulej, K. Comprehensive protocol to simultaneously study protein phosphorylation, acetylation, and N-linked sialylated glycosylation. Methods Mol. Biol. 1295, 275–292 (2015).
Zhang, Y., Wang, H. & Lu, H. Sequential selective enrichment of phosphopeptides and glycopeptides using amine-functionalized magnetic nanoparticles. Mol. Biosyst. 9, 492–500 (2013).
Young, N. L., Plazas-Mayorca, M. D. & Garcia, B. A. Systems-wide proteomic characterization of combinatorial post-translational modification patterns. Expert Rev. Proteom. 7, 79–92 (2010).
Manasanch, E. E. & Orlowski, R. Z. Proteasome inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 14, 417–433 (2017).
Harrigan, J. A., Jacq, X., Martin, N. M. & Jackson, S. P. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat. Rev. Drug Discov. 17, 57–78 (2018).
Soucy, T. A. et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 (2009).
Hyer, M. L. et al. A small-molecule inhibitor of the ubiquitin activating enzyme for cancer treatment. Nat. Med. 24, 186–193 (2018).
He, X. et al. Probing the roles of SUMOylation in cancer cell biology by using a selective SAE inhibitor. Nat. Chem. Biol. 13, 1164–1171 (2017).
Lan, B., Chai, S., Wang, P. & Wang, K. VCP/p97/Cdc48, a linking of protein homeostasis and cancer therapy. Curr. Mol. Med. 17, 608–618 (2017).
Ferguson, F. M. & Gray, N. S. Kinase inhibitors: the road ahead. Nat. Rev. Drug Discov. 17, 353–377 (2018).
Bekker-Jensen, D. B. et al. A compact quadrupole-orbitrap mass spectrometer with FAIMS interface improves proteome coverage in short LC gradients. Mol. Cell Proteom. 19, 716–729 (2020).
Bache, N. et al. A novel LC system embeds analytes in pre-formed gradients for rapid, ultra-robust proteomics. Mol. Cell Proteom. 17, 2284–2296 (2018).
Doll, S., Gnad, F. & Mann, M. The case for proteomics and phospho-proteomics in personalized cancer medicine. Proteom. Clin. Appl. 13, e1800113 (2019).
Meierhofer, D., Wang, X., Huang, L. & Kaiser, P. Quantitative analysis of global ubiquitination in HeLa cells by mass spectrometry. J. Proteome Res. 7, 4566–4576 (2008).
Li, C. et al. Quantitative SUMO proteomics identifies PIAS1 substrates involved in cell migration and motility. Nat. Commun. 11, 834 (2020).
Schimmel, J. et al. Uncovering SUMOylation dynamics during cell-cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Mol. Cell 53, 1053–1066 (2014).
Schimmel, J. et al. The ubiquitin–proteasome system is a key component of the SUMO-2/3 cycle. Mol. Cell Proteom. 7, 2107–2122 (2008).
Tatham, M. H., Matic, I., Mann, M. & Hay, R. T. Comparative proteomic analysis identifies a role for SUMO in protein quality control. Sci. Signal. 4, rs4 (2011).
Vertegaal, A. C. et al. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell Proteom. 5, 2298–2310 (2006).
Li, Z. et al. Functions and substrates of NEDDylation during cell cycle in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 90, 101–112 (2017).
Liao, S., Hu, H., Wang, T., Tu, X. & Li, Z. The protein neddylation pathway in Trypanosoma brucei: functional characterization and substrate identification. J. Biol. Chem. 292, 1081–1091 (2017).
Bennett, E. J., Rush, J., Gygi, S. P. & Harper, J. W. Dynamics of Cullin–RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 143, 951–965 (2010).
Xirodimas, D. P. et al. Ribosomal proteins are targets for the NEDD8 pathway. EMBO Rep. 9, 280–286 (2008).
Jones, J. et al. A targeted proteomic analysis of the ubiquitin-like modifier NEDD8 and associated proteins. J. Proteome Res. 7, 1274–1287 (2008).
Hemelaar, J. et al. Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins. Mol. Cell Biol. 24, 84–95 (2004).
O’Connor, H. F., Swaim, C. D., Canadeo, L. A. & Huibregtse, J. M. Ubiquitin-activated interaction traps (UBAITs): tools for capturing protein–protein interactions. Methods Mol. Biol. 1844, 85–100 (2018).
Shi, Y. et al. A data set of human endogenous protein ubiquitination sites. Mol. Cell Proteom. 10, M110 002089 (2011).
Lopitz-Otsoa, F. et al. SUMO-binding entities (SUBEs) as tools for the enrichment, isolation, identification, and characterization of the SUMO proteome in liver cancer. J. Vis. Exp. https://doi.org/10.3791/60098 (2019).
Bruderer, R. et al. Purification and identification of endogenous polySUMO conjugates. EMBO Rep. 12, 142–148 (2011).
Jeram, S. M. et al. An improved SUMmOn-based methodology for the identification of ubiquitin and ubiquitin-like protein conjugation sites identifies novel ubiquitin-like protein chain linkages. Proteomics 10, 254–265 (2010).
Baldanta, S. et al. ISG15 governs mitochondrial function in macrophages following vaccinia virus infection. PLoS Pathog. 13, e1006651 (2017).
Kozuka-Hata, H. et al. System-wide analysis of protein acetylation and ubiquitination reveals a diversified regulation in human cancer cells. Biomolecules 10, 411 (2020).
Akimov, V. et al. StUbEx PLUS — a modified stable tagged ubiquitin exchange system for peptide level purification and in-depth mapping of ubiquitination sites. J. Proteome Res. 17, 296–304 (2018).
Wagner, S. A. et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell Proteom. 10, M111 013284 (2011).
Hendriks, I. A., D’Souza, R. C., Chang, J. G., Mann, M. & Vertegaal, A. C. System-wide identification of wild-type SUMO-2 conjugation sites. Nat. Commun. 6, 7289 (2015).
Cubenas-Potts, C. et al. Identification of SUMO-2/3-modified proteins associated with mitotic chromosomes. Proteomics 15, 763–772 (2015).
Becker, J. et al. Detecting endogenous SUMO targets in mammalian cells and tissues. Nat. Struct. Mol. Biol. 20, 525–531 (2013).
Matafora, V., D’Amato, A., Mori, S., Blasi, F. & Bachi, A. Proteomics analysis of nucleolar SUMO-1 target proteins upon proteasome inhibition. Mol. Cell Proteom. 8, 2243–2255 (2009).
Giannakopoulos, N. V. et al. Proteomic identification of proteins conjugated to ISG15 in mouse and human cells. Biochem. Biophys. Res. Commun. 336, 496–506 (2005).
Gupta, R. et al. Ubiquitination screen using protein microarrays for comprehensive identification of Rsp5 substrates in yeast. Mol. Syst. Biol. 3, 116 (2007).
Acknowledgements
This work was carried out with financial support from the Natural Sciences and Engineering Research Council (NSERC 311598). IRIC proteomics facility is a Genomics Technology platform funded in part by the Canadian Government through Genome Canada, the Canadian Center of Excellence in Commercialization and Research, and the Canadian Foundation for Innovation. A.C.O.V. is supported by the European Research Council, the Dutch Research Council (NWO) and the Dutch Cancer Society.
Author information
Authors and Affiliations
Contributions
Introduction (P.T.); Experimentation (C.L., T.G.N., P.T. and A.C.O.V.); Results (A.C.O.V.); Applications (P.T.); Reproducibility and data deposition (A.C.O.V.); Limitations and optimizations (A.C.O.V.); Outlook (A.C.O.V.); Figs 1, 2, 3 and 5 (C.L.); Figs 4 and 7 and Box 1 and 2 (C.L. and T.G.N.); Table 1 (C.L., T.G.N. and P.T.); Overview of the Primer (A.C.O.V. and P.T.). All authors reviewed and edited the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
All authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Methods Primers thanks A. Ordureau, J. Peng, A. Pinto-Fernandez, G. Vere and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Biorender: https://biorender.com/
iProX: https://www.iprox.org/
Japan ProteOme Standard Repository/Database: https://jpostdb.org/
JASSA: http://www.jassa.fr
Mass Spectrometry Interactive Virtual Environment (MassIVE): https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp
Panorama Public: https://panoramaweb.org
PeptideAtlas: http://www.peptideatlas.org/
PhosphoSitePlus: https://www.phosphosite.org
PRoteomics IDEntifications Database (PRIDE): https://www.ebi.ac.uk/pride/
UniProt: https://www.uniprot.org
Glossary
- Ubiquitin code
-
The concept that distinct conformations of ubiquitin chains and modifications lead to different cellular outcomes.
- De-NEDDylation
-
Removal of NEDD8 from modified substrates.
- Isopycnic centrifugation
-
A fractionation method where cell components can be separated based on density gradient centrifugation.
- Remnant peptides
-
Amino acids left over on modified lysine residues after proteolytic digestion.
- Ubiquitylome
-
The cell-wide repertoire of ubiquitylated proteins.
- Offline fractionation
-
Fractionation of peptide extracts by methods such as ion exchange or high-pH, reverse-phase chromatography; fractions are subsequently analysed by mass spectrometry.
- Head-to-tail concatemers
-
Long, continuous DNA molecules containing multiple copies of the same gene assembled head to tail.
- UBL traps
-
Affinity purification methods where fusion proteins containing units of ubiquitin-like protein (UBL)-binding domains are expressed in cells to trap UBL conjugates.
- Ubiquitin clipping
-
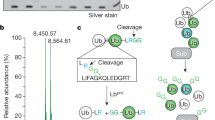
A technique that uses an engineered viral protease, Lbpro∗, to cleave ubiquitin conjugates and leave a traceable diglycine remnant on the modified substrates.
- Pulse chase
-
An experiment where cells are exposed to a labelled compound that is incorporated into proteins, which is later replaced with an unlabelled form to determine the time of exchange.
- Ratio compression
-
Underestimation of the fold-change ratio of peptide/protein abundance in isobaric peptide labelling, owing to co-selection of different peptides during tandem mass spectrometry (MS/MS).
- Translesion synthesis
-
A process whereby the DNA replication machinery can bypass the blocked replication fork caused by DNA damage.
- Affimers
-
Non-antibody binding proteins that mimic the molecular recognition features of antibodies.
- Top-down proteomics
-
A protein identification method that relies on the selection of protein ions as precursors for tandem mass spectrometry (MS/MS) fragmentation.
- Proteolysis-targeting chimaeras
-
(PROTACs). Heterobifunctional small molecules that consist of a ubiquitin E3 ligase-binding domain linked to a domain that bind specifically to a protein targeted for degradation.
Rights and permissions
About this article
Cite this article
Li, C., Nelson, T.G., Vertegaal, A.C.O. et al. Proteomic strategies for characterizing ubiquitin-like modifications. Nat Rev Methods Primers 1, 53 (2021). https://doi.org/10.1038/s43586-021-00048-9
Accepted:
Published:
DOI: https://doi.org/10.1038/s43586-021-00048-9
This article is cited by
-
Deubiquitinating enzymes and the proteasome regulate preferential sets of ubiquitin substrates
Nature Communications (2022)