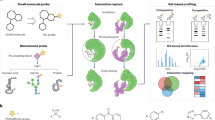
Abstract
Lipids organize the activity of the cell's proteome through a complex network of interactions. The assembly of comprehensive atlases embracing all protein–lipid interactions is an important challenge that requires innovative methods. We recently developed a liposome-microarray-based assay (LiMA) that integrates liposomes, microfluidics and fluorescence microscopy and which is capable of measuring protein recruitment to membranes in a quantitative and high-throughput manner. Compared with previous assays that are labor-intensive and difficult to scale up, LiMA improves the protein–lipid interaction assay throughput by at least three orders of magnitude. Here we provide a step-by-step LiMA protocol that includes the following: (i) the serial and generic production of the liposome microarray; (ii) its integration into a microfluidic format; (iii) the measurement of fluorescently labeled protein (either purified proteins or from cell lysate) recruitment to liposomal membranes using high-throughput microscopy; (iv) automated image analysis pipelines to quantify protein–lipid interactions; and (v) data quality analysis. In addition, we discuss the experimental design, including the relevant quality controls. Overall, the protocol—including device preparation, assay and data analysis—takes 6–8 d. This protocol paves the way for protein–lipid interaction screens to be performed on the proteome and lipidome scales.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
van Meer, G., Voelker, D.R. & Feigenson, G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 (2008).
Scott, J.D. & Pawson, T. Cell signaling in space and time: where proteins come together and when they're apart. Science 326, 1220–1224 (2009).
Leonard, T.A. & Hurley, J.H. Regulation of protein kinases by lipids. Curr. Opin. Struct. Biol. 21, 785–791 (2011).
Fahy, E. et al. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 50 (suppl.), S9–S14 (2009).
Babu, M. et al. Interaction landscape of membrane-protein complexes in Saccharomyces cerevisiae. Nature 489, 585–589 (2012).
Lemmon, M.A. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99–111 (2008).
Wymann, M.P. & Schneiter, R. Lipid signalling in disease. Nat. Rev. Mol. Cell Biol. 9, 162–176 (2008).
Saliba, A.E., Vonkova, I. & Gavin, A.C. The systematic analysis of protein-lipid interactions comes of age. Nat. Rev. Mol Cell Biol. 16, 753–761 (2015).
Lemmon, M.A., Ferguson, K.M., O'Brien, R., Sigler, P.B. & Schlessinger, J. Specific and high-affinity binding of inositol phosphates to an isolated Pleckstrin homology domain. Proc. Natl. Acad. Sci. USA 92, 10472–10476 (1995).
Zhao, H. & Lappalainen, P. A simple guide to biochemical approaches for analyzing protein-lipid interactions. Mol. Biol. Cell 23, 2823–2830 (2012).
Besenicar, M., Macek, P., Lakey, J.H. & Anderluh, G. Surface plasmon resonance in protein-membrane interactions. Chem. Phys. Lipids 141, 169–178 (2006).
Zhu, H. et al. Global analysis of protein activities using proteome chips. Science 293, 2101–2105 (2001).
Haberkant, P. & Holthuis, J.C. Fat & fabulous: bifunctional lipids in the spotlight. Biochim. Biophys. Acta 1841, 1022–1030 (2014).
Maeda, K. et al. Interactome map uncovers phosphatidylserine transport by oxysterol-binding proteins. Nature 501, 257–261 (2013).
Li, X., Gianoulis, T.A., Yip, K.Y., Gerstein, M. & Snyder, M. Extensive in vivo metabolite-protein interactions revealed by large-scale systematic analyses. Cell 143, 639–650 (2010).
Groves, J.T. & Kuriyan, J. Molecular mechanisms in signal transduction at the membrane. Nat. Struct. Mol. Biol. 17, 659–665 (2010).
Ru, H., Zhang, P. & Wu, H. Promiscuity is not always bad. Mol. Cell 54, 208–209 (2014).
Moravcevic, K., Oxley, C.L. & Lemmon, M.A. Conditional peripheral membrane proteins: facing up to limited specificity. Structure 20, 15–27 (2012).
Gallego, O. et al. A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae. Mol. Syst. Biol. 6, 430 (2010).
Vonkova, I. et al. Lipid cooperativity as a general membrane-recruitment principle for PH domains. Cell Rep. 12, 1519–1530 (2015).
Moravcevic, K. et al. Kinase associated-1 domains drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell 143, 966–977 (2010).
Drin, G. Topological regulation of lipid balance in cells. Annu. Rev. Biochem. 83, 51–77 (2014).
Saliba, A.E. et al. A quantitative liposome microarray to systematically characterize protein-lipid interactions. Nat. Methods 11, 47–50 (2014).
Walde, P., Cosentino, K., Engel, H. & Stano, P. Giant vesicles: preparations and applications. Chembiochem 11, 848–865 (2010).
Horger, K.S., Estes, D.J., Capone, R. & Mayer, M. Films of agarose enable rapid formation of giant liposomes in solutions of physiologic ionic strength. J. Am. Chem. Soc. 131, 1810–1819 (2009).
Brehme, M. & Vidal, M. A global protein-lipid interactome map. Mol. Syst. Biol. 6, 443 (2010).
Niphakis, M.J. et al. A global map of lipid-binding proteins and their ligandability in cells. Cell 161, 1668–1680 (2015).
Hoglinger, D., Nadler, A. & Schultz, C. Caged lipids as tools for investigating cellular signaling. Biochim. Biophys. Acta 1841, 1085–1096 (2014).
Carpten, J.D. et al. A transforming mutation in the Pleckstrin homology domain of AKT1 in cancer. Nature 448, 439–444 (2007).
Hammond, G.R., Machner, M.P. & Balla, T. A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J. Cell Biol. 205, 113–126 (2014).
Miao, B. et al. Small molecule inhibition of phosphatidylinositol-3,4,5-triphosphate (PIP3) binding to Pleckstrin homology domains. Proc. Natl. Acad. Sci. USA 107, 20126–20131 (2010).
van den Bogaart, G. et al. Membrane protein sequestering by ionic protein-lipid interactions. Nature 479, 552–555 (2011).
Stefl, M. et al. Dynamics and size of cross-linking-induced lipid nanodomains in model membranes. Biophys. J. 102, 2104–2113 (2012).
Hansen, J.S., Thompson, J.R., Helix-Nielsen, C. & Malmstadt, N. Lipid directed intrinsic membrane protein segregation. J. Am. Chem. Soc. 135, 17294–17297 (2013).
Horger, K.S. et al. Hydrogel-assisted functional reconstitution of human P-glycoprotein (ABCB1) in giant liposomes. Biochim. Biophys. Acta 1848, 643–653 (2015).
Contreras, F.X. et al. Molecular recognition of a single sphingolipid species by a protein's transmembrane domain. Nature 481, 525–529 (2012).
Cesar-Razquin, A. et al. A call for systematic research on solute carriers. Cell 162, 478–487 (2015).
Sampaio, J.L. et al. Membrane lipidome of an epithelial cell line. Proc. Natl. Acad. Sci. USA 108, 1903–1907 (2011).
Tsai, F.C., Stuhrmann, B. & Koenderink, G.H. Encapsulation of active cytoskeletal protein networks in cell-sized liposomes. Langmuir 27, 10061–10071 (2011).
Xia, Y.N. & Whitesides, G.M. Soft lithography. Annu. Rev. Mater. Sci. 28, 153–184 (1998).
Qin, D., Xia, Y. & Whitesides, G.M. Soft lithography for micro- and nanoscale patterning. Nat. Protoc. 5, 491–502 (2010).
Weibel, D.B., Diluzio, W.R. & Whitesides, G.M. Microfabrication meets microbiology. Nat. Rev. Microbiol. 5, 209–218 (2007).
Bartolo, D., Degre, G., Nghe, P. & Studer, V. Microfluidic stickers. Lab Chip 8, 274–279 (2008).
Kim, E., Xia, Y.N. & Whitesides, G.M. Micromolding in capillaries: applications in materials science. J. Am. Chem. Soc. 118, 5722–5731 (1996).
Wu, H.K., Huang, B. & Zare, R.N. Construction of microfluidic chips using polydimethylsiloxane for adhesive bonding. Lab Chip 5, 1393–1398 (2005).
Erfle, H. et al. Reverse transfection on cell arrays for high content screening microscopy. Nat. Protoc. 2, 392–399 (2007).
Neumann, B. et al. High-throughput RNAi screening by time-lapse imaging of live human cells. Nat. Methods 3, 385–390 (2006).
Laufer, C., Fischer, B., Huber, W. & Boutros, M. Measuring genetic interactions in human cells by RNAi and imaging. Nat. Protoc. 9, 2341–2353 (2014).
Rouser, G., Fkeischer, S. & Yamamoto, A. Two dimensional then layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids 5, 494–496 (1970).
Pedelacq, J.D., Cabantous, S., Tran, T., Terwilliger, T.C. & Waldo, G.S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006).
Kotz, K., Cheng, X. & Toner, M. PDMS device fabrication and surface modification. J. Vis. Exp. 8, 319 (2007).
Studier, F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005).
Yamashita, Y., Oka, M., Tanaka, T. & Yamazaki, M. A new method for the preparation of giant liposomes in high salt concentrations and growth of protein microcrystals in them. Biochim. Biophys. Acta 1561, 129–134 (2002).
Duffy, D.C., McDonald, J.C., Schueller, O.J. & Whitesides, G.M. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane). Anal. Chem. 70, 4974–4984 (1998).
Acknowledgements
We are grateful to the EMBL Advanced Light Microscopy Facility (ALMF), C. Gehin and C. Merten for expert help. We also thank other members of P.B.'s, J.E.'s and A.-C.G.'s groups for continuous discussions and support. This work is partially funded by the DFG in the framework of the Cluster of Excellence, CellNetworks Initiative of the University of Heidelberg (ExIni, EcTop). A.-E.S. is supported by the European Molecular Biology Laboratory and the EU Marie Curie Actions Interdisciplinary Postdoctoral Cofunded Programme.
Author information
Authors and Affiliations
Contributions
A.-E.S., A.-C.G. and J.E. designed the research. A.-C.G. directed the research. A.-E.S. developed the experimental protocol with the help of I.V. and S.C. A.-E.S., I.V., K.G.K. and C.T. developed the image analysis pipeline. S.D. and P.B. developed the bioinformatics tools.
Corresponding author
Ethics declarations
Competing interests
A.-E.S., I.V., J.E. and A.-C.G. declare competing financial interests in the form of an international patent application (PCT/EP2013/065256) based on the method LiMA.
Integrated supplementary information
Supplementary Figure 1 Equipment requirement setup.
(a) Glass-slide holder. A home-made glass-slide holder accommodates approx. 20 glass slides with a stirring magnetic bar beneath the glass slides for the washing steps. (b) Spotting rack in a transparent Mylar bag. The spotting rack can be directly plugged in the spotting robot and the lipid solution will be drawn from the vials through the septum. (c) Photo of spotting platform. (d) Photo of the device holder plugged on the microscope.
Supplementary Figure 2 From spotting preparation to spotted-TAL storage.
(a) Photo of the spotting robot. (b-c) After spotting, the protective stickers are removed from the spotted-TALs with tweezers (photo b) and glass slides are removed from the spotting platform (photo c).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–2, Supplementary Data 1 (PDF 518 kb)
Supplementary Table 1
Guide for the calculation of lipid compositions of lipid mixtures used for LiMA (XLSX 17 kb)
Supplementary Data 2
Transparency masks for lithography Raw transparency mask files in dxf format, to make microfluidic channels, spotting platform and protective stickers (ZIP 312 kb)
Supplementary Data 3
CellProfiler pipeline and R script rLiMA (ZIP 37288 kb)
Rights and permissions
About this article
Cite this article
Saliba, AE., Vonkova, I., Deghou, S. et al. A protocol for the systematic and quantitative measurement of protein–lipid interactions using the liposome-microarray-based assay. Nat Protoc 11, 1021–1038 (2016). https://doi.org/10.1038/nprot.2016.059
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.059
This article is cited by
-
Protein–Protein Interactions on Membrane Surfaces Analysed Using Pull-Downs with Supported Bilayers on Silica Beads
The Journal of Membrane Biology (2022)
-
Perturbation of the yeast mitochondrial lipidome and associated membrane proteins following heterologous expression of Artemia-ANT
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.