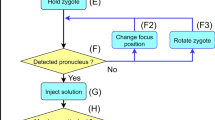
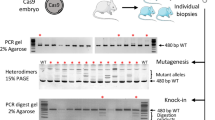
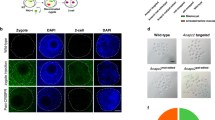
Abstract
Genetically engineered mice are instrumental for the analysis of mammalian gene function in health and disease. As classical gene targeting, which is performed in embryonic stem (ES) cell cultures and generates chimeric mice, is a time-consuming and labor-intensive procedure, we recently used transcription activator–like (TAL) effector nucleases (TALENs) for mutagenesis of the mouse genome directly in one-cell embryos. Here we describe a stepwise protocol for the generation of knock-in and knockout mice, including the selection of TALEN-binding sites, the design and construction of TALEN coding regions and of mutagenic oligodeoxynucleotides (ODNs) and targeting vectors, mRNA production, embryo microinjection and the identification of modified alleles in founder mutants and their progeny. After a setup time of 2–3 weeks of hands-on work for TALEN construction, investigators can obtain first founder mutants for genes of choice within 7 weeks after embryo microinjections.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Capecchi, M.R. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 6, 507–512 (2005).
Rouet, P., Smih, F. & Jasin, M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl. Acad. Sci. USA 91, 6064–6068 (1994).
Porteus, M.H. & Carroll, D. Gene targeting using zinc-finger nucleases. Nat. Biotechnol. 23, 967–973 (2005).
Meyer, M., de Angelis, M.H., Wurst, W. & Kuhn, R. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc. Natl. Acad. Sci. USA 107, 15022–15026 (2010).
Meyer, M., Ortiz, O., Hrabé de Angelis, M., Wurst, W. & Kühn, R. Modeling disease mutations by gene targeting in one-cell mouse embryos. Proc. Natl. Acad. Sci. USA 109, 9354–9359 (2012).
Boch, J. et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512 (2009).
Moscou, M.J. & Bogdanove, A.J. A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501 (2009).
Boch, J. & Bonas, U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol. 48, 419–436 (2010).
Bogdanove, A.J., Schornack, S. & Lahaye, T. TAL effectors: finding plant genes for disease and defense. Curr. Opin. Plant Biol. 13, 394–401 (2010).
Scholze, H. & Boch, J. TAL effectors are remote controls for gene activation. Curr. Opin. Microbiol. 14, 47–53 (2011).
Deng, D. et al. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science 335, 720–723 (2012).
Mak, A.N.-S., Bradley, P., Cernadas, R.A., Bogdanove, A.J. & Stoddard, B.L. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science 335, 716–719 (2012).
Miller, J.C. et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143–148 (2011).
Christian, M. et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186, 757–761 (2010).
Cermak, T. et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82 (2011).
Heyer, W.-D., Ehmsen, K.T. & Liu, J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44, 113–139 (2010).
Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 (2010).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Wang, H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Fu, Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826 (2013).
Hsu, P.D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Wefers, B. et al. Direct production of mouse disease models by embryo microinjection of TALENs and oligodeoxynucleotides. Proc. Natl. Acad. Sci. USA 110, 3782–3787 (2013).
Hockemeyer, D. et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 29, 731–734 (2011).
Wang, H. et al. TALEN-mediated editing of the mouse Y chromosome. Nat. Biotechnol. 31, 530–532 (2013).
Doyle, E.L. et al. TAL effector-nucleotide targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 40, W117–W122 (2012).
Sander, J.D. et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 29, 697–698 (2011).
Meckler, J.F. et al. Quantitative analysis of TALE-DNA interactions suggests polarity effects. Nucleic Acids Res. 41, 4118–4128 (2013).
Streubel, J., Blücher, C., Landgraf, A. & Boch, J. TAL effector RVD specificities and efficiencies. Nat. Biotechnol. 30, 593–595 (2012).
Cong, L., Zhou, R., Kuo, Y.-C., Cunniff, M. & Zhang, F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat. Commun. 3, 968 (2012).
Chen, F. et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods 8, 753–755 (2011).
Wefers, B., Wurst, W. & Kühn, R. Design and Generation of gene-targeting vectors. Curr. Protoc. Mouse Biol. 1, 199–211 (2011).
Hasty, P., Abuin, A. & Bradley, A. Gene targeting, principles, and practice in mammalian cells. in Gene Targeting: A Practical Approach (ed. Joyner, A.L.) 1–35 (Oxford University Press, 2000).
Hasty, P., Rivera-Pérez, J. & Bradley, A. The length of homology required for gene targeting in embryonic stem cells. Mol. Cell Biol. 11, 5586–5591 (1991).
Deng, C. & Capecchi, M.R. Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol. Cell Biol. 12, 3365–3371 (1992).
Reyon, D. et al. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 30, 460–465 (2012).
Briggs, A.W. et al. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res. 40, e117 (2012).
Kim, H. et al. Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nat. Methods 8, 941–943 (2011).
Perez-Pinera, P., Ousterout, D.G., Brown, M.T. & Gersbach, C.A. Gene targeting to the ROSA26 locus directed by engineered zinc-finger nucleases. Nucleic Acids Res. 40, 3741–3752 (2012).
Adenot, P.G., Mercier, Y., Renard, J.P. & Thompson, E.M. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Dev. Camb. Engl. 124, 4615–4625 (1997).
Sung, Y.H. et al. Knockout mice created by TALEN-mediated gene targeting. Nat. Biotechnol. 31, 23–24 (2013).
Tesson, L. et al. Knockout rats generated by embryo microinjection of TALENs. Nat. Biotechnol. 29, 695–696 (2011).
Davies, B. et al. Site-specific mutation of the Zic2 locus by microinjection of TALEN mRNA in mouse CD1, C3H and C57BL/6J oocytes. PLoS ONE 8, e60216 (2013).
Qiu, Z. et al. High-efficiency and heritable gene targeting in mouse by transcription activator-like effector nucleases. Nucleic Acids Res. 41, e120 (2013).
Southern, E. Southern blotting. Nat. Protoc. 1, 518–525 (2006).
Liew, M. et al. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin. Chem. 50, 1156–1164 (2004).
Ittner, L.M. & Götz, J. Pronuclear injection for the production of transgenic mice. Nat. Protoc. 2, 1206–1215 (2007).
Nagy, A., Gertsenstein, M., Vintersten, K. & Behringer, R. Manipulating the Mouse Embryo (Cold Spring Harbour Laboratory Press, 2003).
Sanjana, N.E. et al. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 7, 171–192 (2012).
Schmid-Burgk, J.L., Schmidt, T., Kaiser, V., Höning, K. & Hornung, V. A ligation-independent cloning technique for high-throughput assembly of transcription activator–like effector genes. Nat. Biotechnol. 31, 76–81 (2013).
Acknowledgements
This work was supported by the European Union within the EUCOMMTools project (HEALTH-F4-2010-261492 to W.W.), by the German Ministry of Education and Research within the DIGTOP project (01GS0858 to W.W. and R.K.) of the German National Genome Research Network (NGFN)-Plus program and by the Indian Council of Agricultural Research (no.29-1/2009-EQR/Edn to S.K.P.).
Author information
Authors and Affiliations
Contributions
B.W., S.K.P., O.O., C.B. and R.K. performed the research and analyzed the data; B.W., S.K.P., O.O., C.B., S.H. and R.K. wrote the manuscript; J.H. designed the TALENdesigner tools and webpage; W.W. and R.K. supervised the research.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Genotyping: exemplary results for direct sequencing of PCR products.
Representative chromatograms of heterozygous mutants harboring a single nucleotide substitution (marked in red) (a) or an indel (b) resulting in two superimposed traces.
Supplementary Figure 2 Genotyping: exemplary results of a T7 endonuclease I assay.
Wild-type controls (wt) harbor only the full length PCR product (open triangle), whereas heterozygous mutants (het) show the additional presence of two digestion products (filled triangles) obtained by the cleavage of heteroduplex molecules within the TALEN target region.
Supplementary Figure 3 Genotyping: exemplary results for HRMA analysis.
Representative melting curves of PCR products amplified from a wild-type control (grey) and a mutant founder mouse (red).
Supplementary information
Supplementary Figure 1
Genotyping: exemplary results for direct sequencing of PCR products. (PDF 155 kb)
Supplementary Figure 2
Genotyping: exemplary results of a T7 endonuclease I assay. (PDF 148 kb)
Supplementary Figure 3
Genotyping: exemplary results for HRMA analysis. (PDF 27 kb)
Supplementary Data
(PDF 889 kb)
Pronuclear microinjection
This movie demonstrates the microinjection into the male pronucleus of a mouse one-cell embryo, fixed with a holding pipette. (AVI 2868 kb)
Rights and permissions
About this article
Cite this article
Wefers, B., Panda, S., Ortiz, O. et al. Generation of targeted mouse mutants by embryo microinjection of TALEN mRNA. Nat Protoc 8, 2355–2379 (2013). https://doi.org/10.1038/nprot.2013.142
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2013.142
This article is cited by
-
3R measures in facilities for the production of genetically modified rodents
Lab Animal (2022)
-
Application of CRISPR-Mediated Gene Editing for Crop Improvement
Molecular Biotechnology (2022)
-
ZFP804A mutant mice display sex-dependent schizophrenia-like behaviors
Molecular Psychiatry (2021)
-
The Parkinson’s disease-linked Leucine-rich repeat kinase 2 (LRRK2) is required for insulin-stimulated translocation of GLUT4
Scientific Reports (2019)
-
A history of genome editing in mammals
Mammalian Genome (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.