Abstract
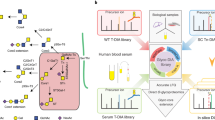
We present a protocol for the identification of glycosylated proteins in plasma followed by elucidation of their individual glycan compositions. The study of glycoproteins by mass spectrometry is usually based on cleavage of glycans followed by separate analysis of glycans and deglycosylated proteins, which limits the ability to derive glycan compositions for individual glycoproteins. The methodology described here consists of 2D HPLC fractionation of intact proteins and liquid chromatography–multistage tandem mass spectrometry (LC-MS/MSn) analysis of digested protein fractions. Protein samples are separated by 1D anion-exchange chromatography (AEX) with an eight-step salt elution. Protein fractions from each of the eight AEX elution steps are transferred onto the 2D reversed-phase column to further separate proteins. A digital ion trap mass spectrometer with a wide mass range is then used for LC-MS/MSn analysis of intact glycopeptides from the 2D HPLC fractions. Both peptide and oligosaccharide compositions are revealed by analysis of the ion fragmentation patterns of glycopeptides with an intact glycopeptide analysis pipeline.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout











Similar content being viewed by others
References
Apweiler, R., Hermjakob, H. & Sharon, N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys Acta. 1473, 4–8 (1999).
Wong, C.H. Protein glycosylation: new challenges and opportunities. J. Org. Chem. 70, 4219–4225 (2005).
Cantagrel, V., Lefeber, D.J., Ng, B.G., Guan, Z., Silhavy, J.L. & Bielas, S.L. et al. SRD5A3 is required for converting polyprenol to dolichol and is mutated in a congenital glycosylation disorder. Cell 142, 203–217 (2010).
Montpetit, M.L., Stocker, P.J., Schwetz, T.A., Harper, J.M., Norring, S.A. & Schaffer, L. et al. Regulated and aberrant glycosylation modulate cardiac electrical signaling. Proc Natl Acad Sci USA 106, 16517–16522 (2009).
Ohtsubo, K. & Marth, J.D. Glycosylation in cellular mechanisms of health and disease. Cell. 126, 855–867 (2006).
Taylor, A.D., Hancock, W.S., Hincapie, M., Taniguchi, N. & Hanash, S.M. Towards an integrated proteomic and glycomic approach to finding cancer biomarkers. Genome Med. 1, 57 (2009).
Peracaula, R. et al. Altered glycosylation pattern allows the distinction between prostate-specific antigen (PSA) from normal and tumor origins. Glycobiology 13, 457–470 (2003).
Leiserowitz, G.S. et al. Glycomics analysis of serum: a potential new biomarker for ovarian cancer? Int. J. Gynecol. Cancer 18, 470–475 (2008).
Hakomori, S. Glycosylation defining cancer malignancy: new wine in an old bottle. Proc. Natl. Acad. Sci. USA 99, 10231–10233 (2002).
Kyselova, Z. et al. Breast cancer diagnosis and prognosis through quantitative measurements of serum glycan profiles. Clin. Chem. 54, 1166–1175 (2008).
Soltermann, A. et al. N-glycoprotein profiling of lung adenocarcinoma pleural effusions by shotgun proteomics. Cancer 114, 124–133 (2008).
Peracaula, R., Barrabés, S., Sarrats, A., Rudd, P.M. & de Llorens, R. Altered glycosylation in tumours focused to cancer diagnosis. Dis. Markers 25, 207–218 (2008).
Gavel, Y. & Von Heijne, G. Sequence difference between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng. 3, 433–442 (1990).
Hansen, J.E. et al. Prediction of O-glycosylation of mammalian proteins: specificity patterns of UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferase. Biochem. J. 308, 801–81 (1995).
Wuhrer, M., Catalina, M.I., Deelder, A.M. & Hokke, C.H. Glycoproteomics based on tandem mass spectrometry of glycopeptides. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 849, 115–128 (2007).
Geyer, H. & Geyer, R. Strategies for analysis of glycoprotein glycosylation. Biochim. Biophys. Acta. 1764, 1853–1869 (2006).
Zhang, H., Li, X.J., Martin, D.B. & Aebersold, R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 21, 660–666 (2003).
Kaji, H. et al. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat. Biotechnol. 21, 667–672 (2003).
Wada, Y. et al. Comparison of the methods for profiling glycoprotein glycans—HUPO Human Disease Glycomics/Proteome Initiative multi-institutional study. Glycobiology 17, 411–422 (2007).
Wuhrer, M., de Boer, A.R. & Deelder, A.M. Structural glycomics using hydrophilic interaction chromatography (HILIC) with mass spectrometry. Mass Spectrom. Rev. 28, 192–206 (2009).
Kirmiz, C. et al. A serum glycomics approach to breast cancer biomarkers. Mol. Cell Proteomics 6, 43–55 (2007).
Li, B. et al. Glycoproteomic analyses of ovarian cancer cell lines and sera from ovarian cancer patients show distinct glycosylation changes in individual proteins. J. Proteome Res. 7, 3776–3788 (2008).
Abd Hamid, U.M. et al. A strategy to reveal potential glycan markers from serum glycoproteins associated with breast cancer progression. Glycobiology 18, 1105–1118 (2008).
Wang, H. & Hanash, S.M. Increased throughput and reduced carryover of mass spectrometry-based proteomics using a high-efficiency nonsplit nanoflow parallel dual-column capillary HPLC system. J. Proteome Res. 7, 2743–2755 (2008).
Faca, V.M. et al. A mouse to human search for plasma proteome changes associated with pancreatic tumor development. PLoS Med. 5, e123 (2008).
Paczesny, S. et al. Elafin is a biomarker of graft-versus-host disease of the skin. Sci. Transl. Med. 2, 13ra2 (2010).
Fukuyama, Y., Nakaya, S., Yamazaki, Y. & Tanaka, K. Ionic liquid matrixes optimized for MALDI-MS of sulfated/sialylated/neutral oligosaccharides and glycopeptides. Anal. Chem. 80, 2171–2179 (2008).
Wang, H. et al. Intact-protein-based high-resolution three-dimensional quantitative analysis system for proteome profiling of biological fluids. Mol. Cell Proteomics 4, 618–625 (2005).
Wang, H. & Hanash, S. Electrospray mass spectrometry for quantitative plasma proteome analysis. Methods Mol. Biol. 564, 227–242 (2009).
Freiwald, A. & Sauer, S. Phylogenetic classification and identification of bacteria by mass spectrometry. Nat. Protoc. 4, 732–742 (2009).
Xu, T., Wong, C.C., Kashina, A. & Yates, J.R. III. Identification of N-terminally arginylated proteins and peptides by mass spectrometry. Nat. Protoc. 4, 325–332 (2009).
Anderson, N.L. & Anderson, N.G. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell Proteomics 1, 845–867 (2002).
Liu, T. et al. Evaluation of multi-protein immunoaffinity subtraction for plasma proteomics and candidate biomarker discovery using mass spectrometry. Mol. Cell Proteomics 5, 2167–2174 (2006).
Polaskova, V., Kapur, A., Khan, A., Molloy, M.P. & Baker, M.S. High-abundance protein depletion: comparison of methods for human plasma biomarker discovery. Electrophoresis 31, 471–482 (2010).
Wada, Y., Tajiri, M. & Yoshida, S. Hydrophilic affinity isolation and MALDI multiple-stage tandem mass spectrometry of glycopeptides for glycoproteomics. Anal. Chem. 76, 6560–6565 (2004).
Sparbier, K., Koch, S., Kessler, I., Wenzel, T. & Kostrzewa, M. Selective isolation of glycoproteins and glycopeptides for MALDI-TOF MS detection supported by magnetic particles. J. Biomol Tech. 16, 407–413 (2005).
Pfenninger, A., Karas, M., Finke, B., Stahl, B. & Sawatzki, G. Matrix optimization for matrix-assisted laser desorption/ionization mass spectrometry of oligosaccharides from human milk. J. Mass Spectrom. 34, 98–104 (1999).
Steven, L., Cohen, S.L. & Chait, B.T. Influence of matrix solution conditions on the MALDI-MS analysis of peptides and proteins. Anal. Chem. 68, 31–37 (1996).
Ding, L., Sudakov, M., Brancia, F.L., Giles, R. & Kumashiro, S. A digital ion trap mass spectrometer coupled with atmospheric pressure ion sources. J. Mass Spectrom. 39, 471–484 (2004).
Craig, R. & Beavis, R.C. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 (2004).
Kersey, P.J. et al. The International Protein Index: an integrated database for proteomics experiments. Proteomics 4, 1985–1988 (2004).
Perkins, D.N., Pappin, D.J., Creasy, D.M. & Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999).
Eng, J.K., McCormack, A.L. & Yates, J.R. III. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994).
Na, S. & Paek, E. Quality assessment of tandem mass spectra based on cumulative intensity normalization. J. Proteome Res. 5, 3241–3248 (2006).
Beausoleil, S.A., Villén, J., Gerber, S.A., Rush, J. & Gygi, S.P. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 (2006).
Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 748, 248–254 (1976).
Nesvizhskii, A.I., Keller, A., Kolker, E. & Aebersold, R.A . Statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 (2003).
Ding, L., Brancia, F.L., Giles, R., Smirnov, S. & Nikolaev, E. Advances of Tandem MS Functions with a Digital Ion Trap. Proc. 53rd ASMS Conf. 5–9 June 2005, San Antonio, Texas.
Baldwin, M.A. et al. Tandem mass spectrometry of peptides with N-terminal glutamine: Studies on a prion protein peptide. J. Am. Soc. Mass Spectrom. 1, 258–264 (1990).
Neta, P., Pu, Q.L., Kilpatrick, L., Yang, X. & Stein, S.E. Dehydration versus deamination of N-terminal glutamine in collision-induced dissociation of protonated peptides. J. Am. Soc. Mass Spectrom. 18, 27–36 (2007).
Author information
Authors and Affiliations
Contributions
H.W., K.T. and S.H. conceived and supervised the project. H.W. contributed to the development and evaluation of the methods and combined the data and wrote the protocol. C.-H.W. designed and developed the analysis software. A.C. performed the sample fractionation. H.W. and S.S. contributed to the mass spectrometry analysis of the sample. H.T. and S.I. optimized the MALDI-DIT instrument. A. Taguchi, A. Taylor and A.C. prepared the analyzed sample. H.T., M.M. and S.K. designed and developed the MALDI-DIT software.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Data 1
Contains input files, MS1 spectra, MS2 spectra and analysis output for Well A10. (ZIP 23419 kb)
Supplementary Data 2
Contains input files, MS1 spectrum, MS2 spectra and analysis output for Well A12. (ZIP 16034 kb)
Supplementary Figure 1
Total ion chromatogram of transferrin tryptic digest analyzed by on-line nano RPLC-ESI-LTQFT. (PPT 87 kb)
Supplementary Figure 2
Isotopic peak distribution for the identified glycopeptide with the sequence R.NEEYN*K.S and the glycan composition 5Hex4HexNAc1Sia. (PPT 141 kb)
Supplementary Table 1
Efficiency of glycopeptides enrichment by hydrophilic interaction chromatography. (DOC 37 kb)
Supplementary Table 2
Protein analysis using nano LC-ESI LTQ-FT. (PDF 51 kb)
Supplementary Table 3
Analysis of glycoproteins in a human plasma fraction by LC MALDI-DIT MS/MS. (PDF 62 kb)
Supplementary Program
Program.zip (21MB) file contains programs, N-glycan database, protein database, refinement databases and readme. (ZIP 21409 kb)
Rights and permissions
About this article
Cite this article
Wang, H., Wong, CH., Chin, A. et al. Integrated mass spectrometry–based analysis of plasma glycoproteins and their glycan modifications. Nat Protoc 6, 253–269 (2011). https://doi.org/10.1038/nprot.2010.176
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2010.176
This article is cited by
-
Structural Studies of Fucosylated N-Glycans by Ion Mobility Mass Spectrometry and Collision-Induced Fragmentation of Negative Ions
Journal of the American Society for Mass Spectrometry (2018)
-
Improvement of mass spectrometry analysis of glycoproteins by MALDI-MS using 3-aminoquinoline/α-cyano-4-hydroxycinnamic acid
Analytical and Bioanalytical Chemistry (2013)
-
Determination of site-specific glycan heterogeneity on glycoproteins
Nature Protocols (2012)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.