Abstract
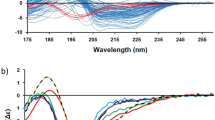
Circular dichroism (CD) is a useful spectroscopic technique for studying the secondary structure, folding and binding properties of proteins. This protocol covers how to use the intrinsic circular dichroic properties of proteins to follow their folding and unfolding as a function of time. Included are methods of obtaining data and for analyzing the folding and unfolding data to determine the rate constants and the order of the folding and unfolding reactions. The protocol focuses on the use of CD to follow folding when it is relatively slow, on the order of minutes to days. The methods for analyzing the data, however, can also be applied to data collected with a CD machine equipped with stopped-flow accessories in the range of milliseconds to seconds and folding analyzed by other spectroscopic methods including changes in absorption or fluorescence spectra as a function of time.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. advance online publication (doi:10.1038/nprot.2006.202).
Greenfield, N.J. Using circular dichroism, collected as a function of temperature, to determine the thermodynamics of protein folding and binding interactions. Nat. Protoc. advance online publication (doi:10.1038/nprot.2006.204).
Greenfield, N.J. Determination of the folding of proteins as a function of denaturants, osmolytes or ligands using circular dichroism. Nat. Protoc. advance online publication (doi:10.1038/nprot.2006.229).
Anfinsen, C.B. & Scheraga, H.A. Experimental and theoretical aspects of protein folding. Adv. Protein. Chem. 29, 205–300 (1975).
Baldwin, R.L. Intermediates in protein folding reactions and the mechanism of protein folding. Annu. Rev. Biochem. 44, 453–475 (1975).
Kuwajima, K. Circular dichroism. Methods Mol. Biol. 40, 115–135 (1995).
Royer, C.A. Fluorescence spectroscopy. Methods Mol. Biol. 40, 65–89 (1995).
Kuwajima, K. & Schmid, F.X. Experimental studies of folding kinetics and structural dynamics of small proteins. Adv. Biophys. 18, 43–74 (1984).
Baldwin, R.L. The nature of protein folding pathways: the classical versus the new view. J. Biomol. NMR 5, 103–109 (1995).
Eftink, M.R. & Shastry, M.C. Fluorescence methods for studying kinetics of protein-folding reactions. Methods Enzymol. 278, 258–286 (1997).
Roder, H., Maki, K., Cheng, H. & Shastry, M.C. Rapid mixing methods for exploring the kinetics of protein folding. Methods 34, 15–27 (2004).
Tanford, C. Physical Chemistry of Macromolecules (John Wiley, New York, 1961).
Dyson, H.J. & Wright, P.E. Insights into protein folding from NMR. Annu. Rev. Phys. Chem. 47, 369–395 (1996).
Dobson, C.M. & Hore, P.J. Kinetic studies of protein folding using NMR spectroscopy. Nat. Struct. Biol. 5 (Suppl): 504–507 (1998).
Konermann, L. & Simmons, D.A. Protein-folding kinetics and mechanisms studied by pulse-labeling and mass spectrometry. Mass Spectrom. Rev. 22, 1–26 (2003).
Krishna, M.M., Hoang, L., Lin, Y. & Englander, S.W. Hydrogen exchange methods to study protein folding. Methods 34, 51–64 (2004).
Konermann, L., Pan, J. & Wilson, D.J. Protein folding mechanisms studied by time-resolved electrospray mass spectrometry. Biotechniques 40, 135–141 (2006).
Piez, K.A. & Sherman, M.R. Equilibrium and kinetic studies of the helix-coil transition in α1-CB2, a small peptide from collagen. Biochemistry 9, 4134–40 (1970).
Wilkinson, R.W. A simple method for determining rate constants and orders of reactions. Chem. Ind. 2, 1395–1397 (1961).
Marquardt, D.W. An algorithm for the estimation of non-linear parameters. J. Soc. Indust. Appl. Math. 11, 431–441 (1963).
Pace, C.N. & McGrath, T. Substrate stabilization of lysozyme to thermal and guanidine hydrochloride denaturation. J. Biol. Chem. 255, 3862–3865 (1980).
Boudko, S. et al. Nucleation and propagation of the collagen triple helix in single-chain and trimerized peptides: transition from third- to first-order kinetics. J. Mol. Biol. 317, 459–470 (2002).
Jackson, S.E. & Fersht, A.R. Folding of chymotrypsin inhibitor 2. 1. Evidence for a two-state transition. Biochemistry 30, 10428–10435 (1991).
Baum, J. & Brodsky, B. Folding of peptide models of collagen and misfolding in disease. Curr. Opin. Struct. Biol. 9, 122–128 (1999).
Sato, S., Luisi, D.L. & Raleigh, D.P. pH jump studies of the folding of the multidomain ribosomal protein L9: the structural organization of the N-terminal domain does not affect the anomalously slow folding of the C-terminal domain. Biochemistry 39, 4955–4962 (2000).
Dobson, C.M. Protein misfolding, evolution and disease. Trends Biochem. Sci. 24, 329–332 (1999).
Hetz, C. & Soto, C. Protein misfolding and disease: the case of prion disorders. Cell. Mol. Life Sci. 60, 133–143 (2003).
Chiti, F. & Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 (2006).
Muchowski, P.J. Protein misfolding, amyloid formation, and neurodegeneration: a critical role for molecular chaperones? Neuron 35, 9–12 (2002).
Melone, M.A., Jori, F.P. & Peluso, G. Huntington's disease: new frontiers for molecular and cell therapy. Curr. Drug Targets 6, 43–56 (2005).
Colon, W. & Kelly, J.W. Partial denaturation of transthyretin is sufficient for amyloid fibril formation in vitro. Biochemistry 31, 8654–8660 (1992).
Hope, J. et al. Cytotoxicity of prion protein peptide (PrP106–126) differs in mechanism from the cytotoxic activity of the Alzheimer's disease amyloid peptide, Aβ25-35. Neurodegeneration 5, 1–11 (1996).
Lai, Z., McCulloch, J., Lashuel, H.A. & Kelly, J.W. Guanidine hydrochloride–induced denaturation and refolding of transthyretin exhibits a marked hysteresis: equilibria with high kinetic barriers. Biochemistry 36, 10230–10239 (1997).
Kapurniotu, A. et al. Contribution of advanced glycosylation to the amyloidogenicity of islet amyloid polypeptide. Eur. J. Biochem. 251, 208–216 (1998).
Kayed, R. et al. Conformational transitions of islet amyloid polypeptide (IAPP) in amyloid formation in vitro. J. Mol. Biol. 287, 781–796 (1999).
Chen, S., Berthelier, V., Yang, W. & Wetzel, R. Polyglutamine aggregation behavior in vitro supports a recruitment mechanism of cytotoxicity. J. Mol. Biol. 311, 173–182 (2001).
Jiang, X. et al. An engineered transthyretin monomer that is nonamyloidogenic, unless it is partially denatured. Biochemistry 40, 11442–11452 (2001).
Zou, W.Q., Yang, D.S., Fraser, P.E., Cashman, N.R. & Chakrabartty, A. All or none fibrillogenesis of a prion peptide. Eur. J. Biochem. 268, 4885–4891 (2001).
Koga, T., Matsuoka, M. & Higashi, N. Structural control of self-assembled nanofibers by artificial β-sheet peptides composed of D- or L-isomer. J. Am. Chem. Soc. 127, 17596–17597 (2005).
Li, J. et al. Structure and influence on stability and activity of the N-terminal propeptide part of lung surfactant protein C. FEBS J. 273, 926–935 (2006).
Hamada, D. & Dobson, C.M. A kinetic study of β-lactoglobulin amyloid fibril formation promoted by urea. Protein Sci. 11, 2417–2426 (2002).
Hortschansky, P., Schroeckh, V., Christopeit, T., Zandomeneghi, G. & Fandrich, M. The aggregation kinetics of Alzheimer's β-amyloid peptide is controlled by stochastic nucleation. Protein Sci. 14, 1753–1759 (2005).
Creighton, T.E. Pathways and mechanisms of protein folding. Adv. Biophys. 18, 1–20 (1984).
Roder, H. & Shastry, M.R. Methods for exploring early events in protein folding. Curr. Opin. Struct. Biol. 9, 620–626 (1999).
Zitzewitz, J.A., Bilsel, O., Luo, J., Jones, B.E. & Matthews, C.R. Probing the folding mechanism of a leucine zipper peptide by stopped-flow circular dichroism spectroscopy. Biochemistry 34, 12812–12819 (1995).
Persikov, A.V., Xu, Y. & Brodsky, B. Equilibrium thermal transitions of collagen model peptides. Protein Sci. 13, 893–902 (2004).
Greenfield, N. et al. Conformational transitions of a collagen fragment studied by CD and 1H-NMR. Biophys. J. 57, 422a (1990).
Piez, K.A. & Sherman, M.R. Characterization of the product formed by renaturation of α1-CB2, a small peptide from collagen. Biochemistry 9, 4129–4133 (1970).
Rossi, A. et al. Type I collagen CNBr peptides: species and behavior in solution. Biochemistry 35, 6048–6057 (1996).
Rossi, A., Zanaboni, G., Cetta, G. & Tenni, R. Stability of type I collagen CNBr peptide trimers. J. Mol. Biol. 269, 488–493 (1997).
Engel, J., Chen, H.T., Prockop, D.J. & Klump, H. The triple helix in equilibrium with coil conversion of collagen-like polytripeptides in aqueous and nonaqueous solvents. Comparison of the thermodynamic parameters and the binding of water to (L-Pro-L-Pro-Gly)n and (L-Pro-L-Hyp-Gly)n. Biopolymers 16, 601–622 (1977).
Long, C.G., Li, M.H., Baum, J. & Brodsky, B. Nuclear magnetic resonance and circular dichroism studies of a triple-helical peptide with a glycine substitution. J. Mol. Biol. 225, 1–4 (1992).
Long, C.G. et al. Characterization of collagen-like peptides containing interruptions in the repeating Gly-X-Y sequence. Biochemistry 32, 11688–11695 (1993).
Bovey, F.A. & Hood, F.P. Circular dichroism spectrum of poly-L-proline. Biopolymers 5, 325–326 (1967).
Greenfield, N. & Fasman, G.D. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry 8, 4108–4116 (1969).
Bentz, H., Bachinger, H.P., Glanville, R. & Kuhn, K. Physical evidence for the assembly of A and B chains of human placental collagen in a single triple helix. Eur. J. Biochem. 92, 563–567 (1978).
Tiffany, M.L. & Krimm, S. Effect of temperature on the circular dichroism spectra of polypeptides in the extended state. Biopolymers 11, 2309–2316 (1972).
Woody, R.W. Circular dichroism and conformation of unordered polypeptides. Adv. Biophys. Chem. 2, 31–79 (1992).
Sreerama, N. & Woody, R.W. Poly(pro)II helices in globular proteins: identification and circular dichroic analysis. Biochemistry 33, 10022–10025 (1994).
Shi, Z., Woody, R.W. & Kallenbach, N.R. Is polyproline II a major backbone conformation in unfolded proteins? Adv. Protein. Chem. 62, 163–240 (2002).
Sato, S., Kuhlman, B., Wu, W.J. & Raleigh, D.P. Folding of the multidomain ribosomal protein L9: the two domains fold independently with remarkably different rates. Biochemistry 38, 5643–5650 (1999).
Acknowledgements
The research was supported by National Institutes of Health grant GM-36326 to N.J.G. and Sarah E. Hitchcock-DeGregori, and by the Circular Dichroism Facility at Robert Wood Johnson Medical School (UMDNJ). In addition, I thank my collaborators and friends, Barbara Brodsky, Gaetano T. Montelione and especially Sarah E. Hitchcock-DeGregori for their encouragement and support of my studies of protein folding and interactions using CD and NMR.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
Supplementary information
Supplementary Equations
Equations for determining rate constants from CD data (PDF 11 kb)
Rights and permissions
About this article
Cite this article
Greenfield, N. Analysis of the kinetics of folding of proteins and peptides using circular dichroism. Nat Protoc 1, 2891–2899 (2006). https://doi.org/10.1038/nprot.2006.244
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.244
This article is cited by
-
Physicochemical and Antioxidant Properties of Industrial Hemp Seed Protein Isolate Treated by High-Intensity Ultrasound
Plant Foods for Human Nutrition (2022)
-
P2R Inhibitors Prevent Antibody-Mediated Complement Activation in an Animal Model of Neuromyelitis Optica
Neurotherapeutics (2022)
-
Higher-Order Structure Comparison of a Proposed Biosimilar and the Innovator Biotherapeutic Trastuzumab using Circular Dichroism Coupled with Statistical Analysis
Journal of Applied Spectroscopy (2020)
-
Immunization with recombinant fusion of LTB and linear epitope (40–62) of epsilon toxin elicits protective immune response against the epsilon toxin of Clostridium perfringens type D
AMB Express (2019)
-
Energy landscapes and functions of supramolecular systems
Nature Materials (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.