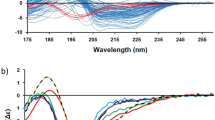
Abstract
Circular dichroism (CD) is an excellent spectroscopic technique for following the unfolding and folding of proteins as a function of temperature. One of its principal applications is to determine the effects of mutations and ligands on protein and polypeptide stability. If the change in CD as a function of temperature is reversible, analysis of the data may be used to determined the van't Hoff enthalpy and entropy of unfolding, the midpoint of the unfolding transition and the free energy of unfolding. Binding constants of protein-protein and protein-ligand interactions may also be estimated from the unfolding curves. Analysis of CD spectra obtained as a function of temperature is also useful to determine whether a protein has unfolding intermediates. Measurement of the spectra of five folded proteins and their unfolding curves at a single wavelength requires ∼8 h.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Velluz, L., Legrand, M. & Grosjean, M. Optical Circular Dichroism: Principles, Measurements, and Applications (Verlag Chemie Academic Press Inc, New York and London, 1965).
Beychok, S. Circular dichroism of biological macromolecules. Science 154, 1288–1299 (1966).
Adler, A.J., Greenfield, N.J. & Fasman, G.D. Circular dichroism and optical rotatory dispersion of proteins and polypeptides. Methods Enzymol. 27, 675–735 (1973).
Johnson, W.C., Jr. Protein secondary structure and circular dichroism: a practical guide. Proteins 7, 205–214 (1990).
Woody, R.W. Circular dichroism. Methods Enzymol. 246, 34–71 (1995).
Kelly, S.M., Jess, T.J. & Price, N.C. How to study proteins by circular dichroism. Biochim. Biophys. Acta 1751, 119–139 (2005).
Miles, A.J. & Wallace, B.A. Synchrotron radiation circular dichroism spectroscopy of proteins and applications in structural and functional genomics. Chem. Soc. Rev. 35, 39–51 (2006).
Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protocols 1, 2006 doi: 10.1038/nprot.2006.202.
Ohgushi, M. & Wada, A. 'Molten-globule state': a compact form of globular proteins with mobile side-chains. FEBS Lett. 164, 21–24 (1983).
Dolgikh, D.A., Abaturov, L.V., Brazhnikov, E.V., Lebedev In, O. & Chirgadze In, N. [Acid form of carbonic anhydrase: “molten globule” with a secondary structure]. Dokl. Akad. Nauk. SSSR 272, 1481–1484 (1983).
Bolotina, I.A. [Secondary structure of proteins from circular dichroism spectra. V. Secondary structure of proteins in a “molten globule” state]. Mol. Biol. (Mosk.) 21, 1625–1635 (1987).
Kuwajima, K. The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins 6, 87–103 (1989).
Goto, Y., Calciano, L.J. & Fink, A.L. Acid-induced folding of proteins. Proc. Natl. Acad. Sci. USA 87, 573–577 (1990).
Greenfield, N.J. Determination of the folding of proteins as a function of denaturants, osmolytes or ligands using circular dichroism. Nat. Protocols 1, 2006 doi: 10.1038/nprot.2006.229.
Greenfield, N.J. Analysis of the kinetics of folding of proteins and peptides using circular dichroism. Nat. Protocols 1, 2006 doi: 10.1038/nprot.2006.244.
Greenfield, N.J. Circular dichroism analysis for protein-protein interactions. Methods Mol. Biol. 261, 55–78 (2004).
Greenfield, N.J. Analysis of circular dichroism data. Methods Enzymol. 383, 282–317 (2004).
Pace, C.N. & McGrath, T. Substrate stabilization of lysozyme to thermal and guanidine hydrochloride denaturation. J. Biol. Chem. 255, 3862–3865 (1980).
Yadav, S. & Ahmad, F. A new method for the determination of stability parameters of proteins from their heat-induced denaturation curves. Anal. Biochem. 283, 207–213 (2000).
Eftink, M.R. Use of multiple spectroscopic methods to monitor equilibrium unfolding of proteins. Methods Enzymol. 259, 487–512 (1995).
Johnson, C.R., Morin, P.E., Arrowsmith, C.H. & Freire, E. Thermodynamic analysis of the structural stability of the tetrameric oligomerization domain of p53 tumor suppressor. Biochemistry 34, 5309–5316 (1995).
Eftink, M.R. et al. Thermodynamics of the unfolding and spectroscopic properties of the V66W mutant of staphylococcal nuclease and its 1–136 fragment. Biochemistry 35, 8084–8094 (1996).
Boice, J.A., Dieckmann, G.R., DeGrado, W.F. & Fairman, R. Thermodynamic analysis of a designed three-stranded coiled coil. Biochemistry 35, 14480–14485 (1996).
Horng, J.C., Moroz, V. & Raleigh, D.P. Rapid cooperative two-state folding of a miniature α-β protein and design of a thermostable variant. J. Mol. Biol. 326, 1261–1270 (2003).
Perczel, A., Hollósi, M., Tusnády, G. & Fasman, G.D. Convex constraint analysis: a natural deconvolution of circular dichroism curves of proteins. Protein Eng. 4, 669–679 (1991).
Greenfield, N.J. & Hitchcock-DeGregori, S.E. Conformational intermediates in the folding of a coiled-coil model peptide of the N-terminus of tropomyosin and α α-tropomyosin. Protein Sci. 2, 1263–1273 (1993).
Safar, J., Roller, P.P., Gajdusek, D.C. & Gibbs, C.J., Jr. Thermal stability and conformational transitions of scrapie amyloid (prion) protein correlate with infectivity. Protein Sci. 2, 2206–2216 (1993).
Staskus, P.W. & Johnson, W.C., Jr. Conformational transition of hyaluronic acid in aqueous-organic solvent monitored by vacuum ultraviolet circular dichroism. Biochemistry 27, 1522–1527 (1988).
McPhie, P. & Shrager, R.I. An investigation of the thermal unfolding of swine pepsinogen using circular dichroism. Arch. Biochem. Biophys. 293, 46–53 (1992).
Konno, T. Conformational diversity of acid-denatured cytochrome c studied by a matrix analysis of far-UV CD spectra. Protein Sci. 7, 975–982 (1998).
Ionescu, R.M., Smith, V.F., O'Neill, J.C., Jr. & Matthews, C.R. Multistate equilibrium unfolding of Escherichia coli dihydrofolate reductase: thermodynamic and spectroscopic description of the native, intermediate, and unfolded ensembles. Biochemistry 39, 9540–9550 (2000).
Thompson, K.S., Vinson, C.R. & Freire, E. Thermodynamic characterization of the structural stability of the coiled-coil region of the bZIP transcription factor GCN4. Biochemistry 32, 5491–5496 (1993).
Taylor, J.W., Greenfield, N.J., Wu, B. & Privalov, P.L. A calorimetric study of the folding-unfolding of an α-helix with covalently closed N and C-terminal loops. J. Mol. Biol. 291, 965–976 (1999).
Singh, A. & Hitchcock-DeGregori, S.E. Local destabilization of the tropomyosin coiled coil gives the molecular flexibility required for actin binding. Biochemistry 42, 14114–14121 (2003).
Ramsay, G.D. & Eftink, M.R. Analysis of multidimensional spectroscopic data to monitor unfolding of proteins. Methods Enzymol. 240, 615–645 (1994).
Schellman, J.A. Macromolecular binding. Biopolymers 14, 999–1018 (1975).
Schellman, J.A. The effect of binding on the melting temperature of biopolymers. Biopolymers 15, 999–1000 (1976).
Marquardt, D.W. An algorithm for the estimation of non-linear parameters. J. Soc. Indust. Appl. Math. 11, 431–441 (1963).
Greenfield, N.J., Montelione, G.T., Farid, R.S. & Hitchcock-DeGregori, S.E. The structure of the N-terminus of striated muscle α-tropomyosin in a chimeric peptide: nuclear magnetic resonance structure and circular dichroism studies. Biochemistry 37, 7834–7843 (1998).
Greenfield, N.J., Palm, T. & Hitchcock-DeGregori, S.E. Structure and interactions of the carboxyl terminus of striated muscle α-tropomyosin: it is important to be flexible. Biophys. J. 83, 2754–2766 (2002).
Greenfield, N.J. et al. The structure of the carboxyl terminus of striated α-tropomyosin in solution reveals an unusual parallel arrangement of interacting α-helices. Biochemistry 42, 614–619 (2003).
Palm, T., Greenfield, N.J. & Hitchcock-DeGregori, S.E. Tropomyosin ends determine the stability and functionality of overlap and troponin T complexes. Biophys. J. 84, 3181–3189 (2003).
Palm, T., Graboski, S., Hitchcock-DeGregori, S.E. & Greenfield, N.J. Disease-causing mutations in cardiac troponin T: identification of a critical tropomyosin-binding region. Biophys. J. 81, 2827–2837 (2001).
Greenfield, N. & Fasman, G.D. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry 8, 4108–4116 (1969).
Bentz, H., Bächinger, H.P., Glanville, R. & Kühn, K. Physical evidence for the assembly of A and B chains of human placental collagen in a single triple helix. Eur. J. Biochem. 92, 563–567 (1978).
Tiffany, M.L. & Krimm, S. Effect of temperature on the circular dichroism spectra of polypeptides in the extended state. Biopolymers 11, 2309–2316 (1972).
Woody, R.W. Circular dichroism and conformation of unordered polypeptides. Adv. Biophys. Chem. 2, 31–79 (1992).
Acknowledgements
The work was supported by a National Institutes of Health grant, GM-36326 to N.J.G. and Dr. Sarah E. Hitchcock-DeGregori, and by the Circular Dichroism Facility at Robert Wood Johnson Medical School (UMDNJ). I thank the late Dr. Gerald D. Fasman for supplying copies of programs to deconvolute data sets using the CCA and Dr. Takashi Konno for supplying a SVD program.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
Supplementary information
Supplementary Equations
Equations for determining the midpoint of a unfolding or folding transition, TM, and enthalpy of folding, ΔH, from changes in ellipticity as a function of temperature. (PDF 45 kb)
Supplementary Tutorial
Deconvolution of a set of curves with the convex constraint algorithm. (PDF 26 kb)
Supplementary Program
DOS version of the CCA for deconvoluting sets of CD spectra. (ZIP 101 kb)
Supplementary Program (use if having trouble unzipping Supplementary Program above)
DOS version of the CCA for deconvoluting sets of CD spectra. (EXE 148 kb)
Rights and permissions
About this article
Cite this article
Greenfield, N. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat Protoc 1, 2527–2535 (2006). https://doi.org/10.1038/nprot.2006.204
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.204
This article is cited by
-
The structural characterization and bioactivity assessment of nonspecific lipid transfer protein 1 (nsLTP1) from caraway (Carum carvi) seeds
BMC Complementary Medicine and Therapies (2023)
-
Design of allosteric sites into rotary motor V1-ATPase by restoring lost function of pseudo-active sites
Nature Chemistry (2023)
-
Heating-mediated purification of active FGF21 and structure-based design of its variant with enhanced potency
Scientific Reports (2023)
-
Structure and mechanism of oxalate transporter OxlT in an oxalate-degrading bacterium in the gut microbiota
Nature Communications (2023)
-
cDNA Cloning, Heterologous Expression, Cytotoxicity, and Inhibitory Effects of a Disintegrin from Bothrops ammodytoides Venom
International Journal of Peptide Research and Therapeutics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.