Abstract
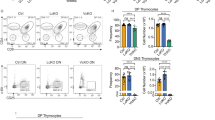
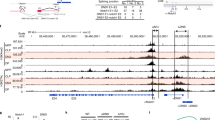
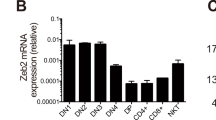
The products of recombination-activating gene 1 (Rag1) and Rag2 are required for T cell receptor gene assembly and thymocyte maturation, yet their transcriptional control mechanisms remain unclear. A congenic strain (called 'ZORI' here) with defects in Rag1 and Rag2 expression, thymocyte maturation and peripheral T cell homeostasis has been developed. Here, we mapped the mutation in this strain to a chromosome 18 locus containing a single known gene encoding the zinc finger protein Zfp608. This gene (Zfp608) was highly expressed in neonatal thymus but was extinguished thereafter. In contrast to wild-type mice, ZORI mice had sustained thymocyte expression of Zfp608 throughout life. The ZORI mutation produced a thymocyte-intrinsic developmental defect. Overexpression of Zfp608 in BALB/c thymocytes substantially impaired Rag1 and Rag2 expression, indicating the underlying mechanism for the defect in ZORI thymocyte development. Thus, the normal function of Zfp608 may be to prevent Rag1 and Rag2 expression in utero.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Godfrey, D.I., Kennedy, J., Suda, T. & Zlotnik, A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J. Immunol. 150, 4244–4252 (1993).
Mombaerts, P. et al. Mutations in T-cell antigen receptor genes α and β block thymocyte development at different stages. Nature 360, 225–231 (1992).
Malissen, M. et al. T cell development in mice lacking the CD3-ζ/ε gene. EMBO J. 12, 4347–4355 (1993).
Fehling, H.J., Krotkova, A., Saint-Ruf, C. & von Boehmer, H. Crucial role of the pre-T-cell receptor α gene in development of αβ but not γδ T cells. Nature 375, 795–798 (1995).
Malissen, M. et al. Regulation of TCR α and β gene allelic exclusion during T-cell development. Immunol. Today 13, 315–322 (1992).
McBlane, J.F. et al. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell 83, 387–395 (1995).
Schatz, D.G., Oettinger, M.A. & Baltimore, D. The V(D)J recombination activating gene, RAG-1. Cell 59, 1035–1048 (1989).
Wilson, A., Held, W. & MacDonald, H.R. Two waves of recombinase gene expression in developing thymocytes. J. Exp. Med. 179, 1355–1360 (1994).
Taniguchi, M., Harada, M., Kojo, S., Nakayama, T. & Wakao, H. The regulatory role of Vα14 NKT cells in innate and acquired immune response. Annu. Rev. Immunol. 21, 483–513 (2003).
Chun, T. et al. CD1d-expressing dendritic cells but not thymic epithelial cells can mediate negative selection of NKT cells. J. Exp. Med. 197, 907–918 (2003).
Zhang, F. et al. A murine locus on chromosome 18 controls NKT cell homeostasis and Th cell differentiation. J. Immunol. 171, 4613–4620 (2003).
Cipollina, C., Alberghina, L., Porro, D. & Vai, M. SFP1 is involved in cell size modulation in respiro-fermentative growth conditions. Yeast 22, 385–399 (2005).
Fingerman, I., Nagaraj, V., Norris, D. & Vershon, A.K. Sfp1 plays a key role in yeast ribosome biogenesis. Eukaryot. Cell 2, 1061–1068 (2003).
Jorgensen, P., Nishikawa, J.L., Breitkreutz, B.J. & Tyers, M. Systematic identification of pathways that couple cell growth and division in yeast. Science 297, 395–400 (2002).
Marion, R.M. et al. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl. Acad. Sci. USA 101, 14315–14322 (2004).
Xu, Z. & Norris, D. The SFP1 gene product of Saccharomyces cerevisiae regulates G2/M transitions during the mitotic cell cycle and DNA-damage response. Genetics 150, 1419–1428 (1998).
Blumberg, H. & Silver, P. A split zinc-finger protein is required for normal yeast growth. Gene 107, 101–110 (1991).
Deftos, M.L. & Bevan, M.J. Notch signaling in T cell development. Curr. Opin. Immunol. 12, 166–172 (2000).
Deftos, M.L., He, Y.W., Ojala, E.W. & Bevan, M.J. Correlating notch signaling with thymocyte maturation. Immunity 9, 777–786 (1998).
Hoebeke, I. et al. Overexpression of HES-1 is not sufficient to impose T-cell differentiation on human hematopoietic stem cells. Blood 107, 2879–2881 (2006).
Kawamata, S., Du, C., Li, K. & Lavau, C. Overexpression of the Notch target genes Hes in vivo induces lymphoid and myeloid alterations. Oncogene 21, 3855–3863 (2002).
Agrawal, A., Eastman, Q.M. & Schatz, D.G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394, 744–751 (1998).
Hiom, K., Melek, M. & Gellert, M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell 94, 463–470 (1998).
Zarrin, A.A., Fong, I., Malkin, L., Marsden, P.A. & Berinstein, N.L. Cloning and characterization of the human recombination activating gene 1 (RAG1) and RAG2 promoter regions. J. Immunol. 159, 4382–4394 (1997).
Kurioka, H. et al. Isolation and characterization of a TATA-less promoter for the human RAG-1 gene. Mol. Immunol. 33, 1059–1066 (1996).
Fuller, K. & Storb, U. Identification and characterization of the murine Rag1 promoter. Mol. Immunol. 34, 939–954 (1997).
Lauring, J. & Schlissel, M.S. Distinct factors regulate the murine Rag-2 promoter in B- and T-cell lines. Mol. Cell. Biol. 19, 2601–2612 (1999).
Kishi, H. et al. Cooperative binding of c-Myb and Pax-5 activates the Rag-2 promoter in immature B cells. Blood 99, 576–583 (2002).
Kishi, H. et al. Lineage-specific regulation of the murine Rag-2 promoter: GATA-3 in T cells and Pax-5 in B cells. Blood 95, 3845–3852 (2000).
Jin, Z.X. et al. Lymphoid enhancer-binding factor-1 binds and activates the recombination-activating gene-2 promoter together with c-Myb and Pax-5 in immature B cells. J. Immunol. 169, 3783–3792 (2002).
Brown, S.T. et al. Regulation of the RAG-1 promoter by the NF-Y transcription factor. J. Immunol. 158, 5071–5074 (1997).
Fong, I.C., Zarrin, A.A., Wu, G.E. & Berinstein, N.L. Functional analysis of the human RAG 2 promoter. Mol. Immunol. 37, 391–402 (2000).
Wang, Q.F., Lauring, J. & Schlissel, M.S. c-Myb binds to a sequence in the proximal region of the Rag-2 promoter and is essential for promoter activity in T-lineage cells. Mol. Cell. Biol. 20, 9203–9211 (2000).
Hsu, L.Y. et al. A conserved transcriptional enhancer regulates RAG gene expression in developing B cells. Immunity 19, 105–117 (2003).
Yu, W. et al. Coordinate regulation of RAG1 and RAG2 by cell type-specific DNA elements 5′ of RAG2. Science 285, 1080–1084 (1999).
Monroe, R.J., Chen, F., Ferrini, R., Davidson, L. & Alt, F.W. RAG2 is regulated differentially in B and T cells by elements 5′ of the promoter. Proc. Natl. Acad. Sci. USA 96, 12713–12718 (1999).
Wei, X.C. et al. Characterization of chromatin structure and enhancer elements for murine recombination activating gene-2. J. Immunol. 169, 873–881 (2002).
Dudley, E.C., Petrie, H.T., Shah, L.M., Owen, M.J. & Hayday, A.C. T cell receptor beta chain gene rearrangement and selection during thymocyte development in adult mice. Immunity 1, 83–93 (1994).
Mombaerts, P. et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68, 869–877 (1992).
Shinkai, Y. et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 68, 855–867 (1992).
Piguet, P.F., Irle, C., Kollatte, E. & Vassalli, P. Post-thymic T lymphocyte maturation during ontogenesis. J. Exp. Med. 154, 581–593 (1981).
Spear, P.G., Wang, A.L., Rutishauser, U. & Edelman, G.M. Characterization of splenic lymphoid cells in fetal and newborn mice. J. Exp. Med. 138, 557–573 (1973).
Spear, P.G. & Edelman, G.M. Maturation of the humoral immune response in mice. J. Exp. Med. 139, 249–263 (1974).
Amagai, T., Itoi, M. & Kondo, Y. Limited development capacity of the earliest embryonic murine thymus. Eur. J. Immunol. 25, 757–762 (1995).
Schlissel, M.S., Corcoran, L.M. & Baltimore, D. Virus-transformed pre-B cells show ordered activation but not inactivation of immunoglobulin gene rearrangement and transcription. J. Exp. Med. 173, 711–720 (1991).
Whitehurst, C.E., Chattopadhyay, S. & Chen, J. Control of V(D)J recombinational accessibility of the Dβ1 gene segment at the TCRβ locus by a germline promoter. Immunity 10, 313–322 (1999).
Goldman, J.P., Spencer, D.M. & Raulet, D.H. Ordered rearrangement of variable region genes of the T cell receptor γ locus correlates with transcription of the unrearranged genes. J. Exp. Med. 177, 729–739 (1993).
Capone, M., Hockett, R.D., Jr. & Zlotnik, A. Kinetics of T cell receptor β, γ, and δ rearrangements during adult thymic development: T cell receptor rearrangements are present in CD44+CD25+ pro-T thymocytes. Proc. Natl. Acad. Sci. USA 95, 12522–12527 (1998).
Sikes, M.L., Gomez, R.J., Song, J. & Oltz, E.M. A developmental stage-specific promoter directs germline transcription of Dβ Jβ gene segments in precursor T lymphocytes. J. Immunol. 161, 1399–1405 (1998).
Acknowledgements
We thank J.W. Thomas and L. Van Kaer for comments and review of the manuscript, and J. Sligh for comments and assistance with the temperature-gradient capillary electrophoresis technique; M. Boothby (Vanderbilt University) provided OTII OVA-specific TCR-transgenic mice. Supported by the US National Institutes of Health (5 T32 DK07563 to F.Z., AI 44924 to T.M.A., P01 HL68744 and CA100905 to E.M.O., and F32 AI066691 and 5T32 HL069765 to L.R.T.).
Author information
Authors and Affiliations
Contributions
All authors contributed to discussions, experimental design, data analysis and preparation of the manuscript; F.Z. designed and executed experiments and prepared the manuscript; L.R.T. examined TCR rearrangement and helped to analyze data and to write the manuscript; and T.M.A. and E.M.O. supervised the work and assisted in data analysis and composition of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
T cell development defects in the ZORI stain map to a locus between 54.3 Mb (D18mit150) and 55.8 Mb (D18mit218) on chromosome 18. (PDF 307 kb)
Supplementary Fig. 2
Relative expression of genes adjacent to Zfp608 on chromosome 18. (PDF 260 kb)
Supplementary Note 1
MapPair primer sequences and primer sequences for TCR rearrangements. (PDF 7 kb)
Rights and permissions
About this article
Cite this article
Zhang, F., Thomas, L., Oltz, E. et al. Control of thymocyte development and recombination-activating gene expression by the zinc finger protein Zfp608. Nat Immunol 7, 1309–1316 (2006). https://doi.org/10.1038/ni1397
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni1397
This article is cited by
-
Reciprocal regulation of Rag expression in thymocytes by the zinc-finger proteins, Zfp608 and Zfp609
Genes & Immunity (2013)
-
Fetal thymocyte development: putting a zinc finger on it?
Nature Immunology (2006)