Abstract
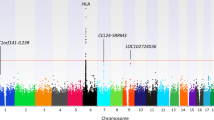
Single-nucleotide variations in C13orf31 (LACC1) that encode p.C284R and p.I254V in a protein of unknown function (called 'FAMIN' here) are associated with increased risk for systemic juvenile idiopathic arthritis, leprosy and Crohn's disease. Here we set out to identify the biological mechanism affected by these coding variations. FAMIN formed a complex with fatty acid synthase (FASN) on peroxisomes and promoted flux through de novo lipogenesis to concomitantly drive high levels of fatty-acid oxidation (FAO) and glycolysis and, consequently, ATP regeneration. FAMIN-dependent FAO controlled inflammasome activation, mitochondrial and NADPH-oxidase-dependent production of reactive oxygen species (ROS), and the bactericidal activity of macrophages. As p.I254V and p.C284R resulted in diminished function and loss of function, respectively, FAMIN determined resilience to endotoxin shock. Thus, we have identified a central regulator of the metabolic function and bioenergetic state of macrophages that is under evolutionary selection and determines the risk of inflammatory and infectious disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Wakil, S.M. et al. Association of a mutation in LACC1 with a monogenic form of systemic juvenile idiopathic arthritis. Arthritis Rheumatol. 67, 288–295 (2015).
Patel, N. et al. Study of Mendelian forms of Crohn's disease in Saudi Arabia reveals novel risk loci and alleles. Gut 63, 1831–1832 (2014).
Liu, H. et al. Discovery of six new susceptibility loci and analysis of pleiotropic effects in leprosy. Nat. Genet. 47, 267–271 (2015).
Jostins, L. et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012).
Hruz, T. et al. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics 2008, 420747 (2008).
Heng, T.S. & Painter, M.W. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008).
Jensen-Urstad, A.P. & Semenkovich, C.F. Fatty acid synthase and liver triglyceride metabolism: housekeeper or messenger? Biochim. Biophys. Acta 1821, 747–753 (2012).
Jensen-Urstad, A.P. et al. Nutrient-dependent phosphorylation channels lipid synthesis to regulate PPARα. J. Lipid Res. 54, 1848–1859 (2013).
Lodhi, I.J. & Semenkovich, C.F. Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab. 19, 380–392 (2014).
Murray, P.J. et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41, 14–20 (2014).
Tannahill, G.M. et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496, 238–242 (2013).
Huang, S.C. et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat. Immunol. 15, 846–855 (2014).
Skarnes, W.C. et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 (2011).
Hillebrand, M. et al. Identification of a new fatty acid synthesis-transport machinery at the peroxisomal membrane. J. Biol. Chem. 287, 210–221 (2012).
Semenkovich, C.F. Regulation of fatty acid synthase (FAS). Prog. Lipid Res. 36, 43–53 (1997).
Jha, A.K. et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–430 (2015).
Grevengoed, T.J., Klett, E.L. & Coleman, R.A. Acyl-CoA metabolism and partitioning. Annu. Rev. Nutr. 34, 1–30 (2014).
Carracedo, A., Cantley, L.C. & Pandolfi, P.P. Cancer metabolism: fatty acid oxidation in the limelight. Nat. Rev. Cancer 13, 227–232 (2013).
Wu, M. et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am. J. Physiol. Cell Physiol. 292, C125–C136 (2007).
Hao, W., Chang, C.P., Tsao, C.C. & Xu, J. Oligomycin-induced bioenergetic adaptation in cancer cells with heterogeneous bioenergetic organization. J. Biol. Chem. 285, 12647–12654 (2010).
Newsholme, E.A., Sugden, P.H. & Williams, T. Effect of citrate on the activities of 6-phosphofructokinase from nervous and muscle tissues from different animals and its relationships to the regulation of glycolysis. Biochem. J. 166, 123–129 (1977).
Fantin, V.R., St-Pierre, J. & Leder, P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 9, 425–434 (2006).
Vats, D. et al. Oxidative metabolism and PGC-1β attenuate macrophage-mediated inflammation. Cell Metab. 4, 13–24 (2006).
Landree, L.E. et al. C75, a fatty acid synthase inhibitor, modulates AMP-activated protein kinase to alter neuronal energy metabolism. J. Biol. Chem. 279, 3817–3827 (2004).
Liesa, M. & Shirihai, O.S. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 17, 491–506 (2013).
Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 417, 1–13 (2009).
Yu, L., Quinn, M.T., Cross, A.R. & Dinauer, M.C. Gp91phox is the heme binding subunit of the superoxide-generating NADPH oxidase. Proc. Natl. Acad. Sci. USA 95, 7993–7998 (1998).
Ran, F.A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
West, A.P. et al. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature 472, 476–480 (2011).
Kampmann, B. et al. Evaluation of human antimycobacterial immunity using recombinant reporter mycobacteria. J. Infect. Dis. 182, 895–901 (2000).
Zhou, R., Yazdi, A.S., Menu, P. & Tschopp, J. A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225 (2011).
Gattorno, M. et al. The pattern of response to anti-interleukin-1 treatment distinguishes two subsets of patients with systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. 58, 1505–1515 (2008).
Mellins, E.D., Macaubas, C. & Grom, A.A. Pathogenesis of systemic juvenile idiopathic arthritis: some answers, more questions. Nat. Rev. Rheumatol. 7, 416–426 (2011).
Quartier, P. et al. A multicentre, randomised, double-blind, placebo-controlled trial with the interleukin-1 receptor antagonist anakinra in patients with systemic-onset juvenile idiopathic arthritis (ANAJIS trial). Ann. Rheum. Dis. 70, 747–754 (2011).
Pascual, V., Allantaz, F., Arce, E., Punaro, M. & Banchereau, J. Role of interleukin-1 (IL-1) in the pathogenesis of systemic onset juvenile idiopathic arthritis and clinical response to IL-1 blockade. J. Exp. Med. 201, 1479–1486 (2005).
Leist, M., Single, B., Castoldi, A.F., Kühnle, S. & Nicotera, P. Intracellular adenosine triphosphate (ATP) concentration: a switch in the decision between apoptosis and necrosis. J. Exp. Med. 185, 1481–1486 (1997).
Hue, L. & Taegtmeyer, H. The Randle cycle revisited: a new head for an old hat. Am. J. Physiol. Endocrinol. Metab. 297, E578–E591 (2009).
Guan, H.P. et al. A futile metabolic cycle activated in adipocytes by antidiabetic agents. Nat. Med. 8, 1122–1128 (2002).
O'Sullivan, D. et al. Memory CD8+ T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity 41, 75–88 (2014).
van der Windt, G.J. et al. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proc. Natl. Acad. Sci. USA 110, 14336–14341 (2013).
Beloqui, A. et al. Novel polyphenol oxidase mined from a metagenome expression library of bovine rumen: biochemical properties, structural analysis, and phylogenetic relationships. J. Biol. Chem. 281, 22933–22942 (2006).
Kim, Y. et al. Crystal structure of hypothetical protein YfiH from Shigella flexneri at 2 A resolution. Proteins 63, 1097–1101 (2006).
Samudio, I. et al. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J. Clin. Invest. 120, 142–156 (2010).
Berod, L. et al. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat. Med. 20, 1327–1333 (2014).
Hall, C.J. et al. Immunoresponsive gene 1 augments bactericidal activity of macrophage-lineage cells by regulating β-oxidation-dependent mitochondrial ROS production. Cell Metab. 18, 265–278 (2013).
O'Neill, L.A. & Pearce, E.J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 213, 15–23 (2016).
Mulders-Manders, C.M. & Simon, A. Hyper-IgD syndrome/mevalonate kinase deficiency: what is new? Semin. Immunopathol. 37, 371–376 (2015).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Behringer, R., Gertsenstein, M., Vintersten Nagy, K. & Nagy, A. Manipulating the Mouse Embryo: A Laboratory Manual 4th edn. 92–93, 139–142 and 211–215 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2014).
Bougaki, M. et al. Nos3 protects against systemic inflammation and myocardial dysfunction in murine polymicrobial sepsis. Shock 34, 281–290 (2010).
Acknowledgements
We thank G. Brown for help with laccase assays; J. Murkin and M. Deery for proteomics; L. Porter for metabolic-flux assays; J. Skepper for electron microscopy; I. Purvis for help with in vivo procedures; R. Rodrigues, M. Md-Ibrahim and J. Jones for cellular assays; S. Dhillon for the generation of constructs; T. Lawley, M. Pardo, J. Choudhary, K. Smith, J. Lee, D. Thomas, G. Schneditz, L. Haag, M. Parkes and R. Blumberg for discussions; all National Institute for Health Research Cambridge BioResource volunteers for the participation; the Cambridge BioResource staff for help with volunteer recruitment; members of the Cambridge BioResource SAB and Management Committee for support of this study; and the National Institute for Health Research Cambridge BRC Cell Phenotyping Hub for expertise and help. Access to Cambridge BioResource volunteers and their data and samples is governed by the Cambridge BioResource SAB (documents on access arrangements and contact details, http://www.cambridgebioresource.org.uk/). Supported by the European Research Council under the European Community's Seventh Framework Programme (FP7/2007-2013)/ERC Grant agreement 260961 (A.K.), the Wellcome Trust (investigator award 106260/Z/14/Z to A.K.; a PhD fellowship for clinicians to M.Z.C.; and a Career Re-Entry Fellowship to N.C.K.), the Wellcome Trust Sanger Institute (G.D., A.B., S.M., S.C. and K.B.), the US National Institutes of Health (5U420D011174 and 5U54HG006348 to A.B. and K.B.), the Biotechnology and Biological Sciences Research Council (M.J.O.W.), the National Institute for Health Research Cambridge Biomedical Research Centre, the European Crohn's and Colitis Organisation (M.T.) and the Swedish Medical Research Council and the Olle Engkvist foundation (M.D'A.).
Author information
Authors and Affiliations
Contributions
M.Z.C., together with S.L.K., G.W.S., S.S., J.W.A., M.T., T.R. and N.C.K., designed and performed most of the experiments; K.B., B.D. and A.B. designed, generated and confirmed the genotype of CRISPR-Cas9–generated mouse lines; Q.Z. and M.J.O.W. provided lipidomics experiments and analysis; G.A. and M.D'A. identified the cellular localization of FAMIN; S.C., S.M. and G.D. contributed Salmonella and part of the in vivo experimentation; K.P.B. and R.A.F. contributed to mycobacterial experiments; E.R.C. helped with metabolic-flux assays and ROS experimentation; J.L.G. contributed metabolomics experimentation and analysis; and A.K. devised and coordinated the project and, together with M.Z.C. and G.D., and with contributions from all authors, designed experiments, interpreted data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Expression of Strep-tagged FAMIN in HEK293T cells.
Immunoblots (IB) of HEK293T lysates expressing N- and C-terminally Strep-tagged FAMIN(p.254I) and FAMIN(p.254V) variants for FAMIN and Strep-tag; β-actin loading control. Data are representative of three independent experiments.
Supplementary Figure 2 FAMIN localizes to peroxisomes.
(a) Co-localization by immunofluorescence (IF) of FAMIN (red) with PMP70 (green) in primary human macrophages (MΦ). DAPI nuclear staining, blue; scale bar = 10 μm. White box corresponds to enlarged images shown in Fig. 1d. (b–e) Co-localization by IF of FAMIN (red) with PMP70 (green), catalase (green), cytochrome oxidase IV (green) and calreticulin (green) in THP-1 macrophages. DAPI, blue; scale bar = 10 μm for original images and 5 μm for enlarged images. Upper panels: original image, middle panels: enlargement of the area shown in the white box, lower panels: further enlargement as indicated. No significant co-localization of FAMIN was detected with CENP-A, centromere; caveolin-2, cholesterol/sphingolipid enriched plasma membrane; EEA1 and RAB5, early endosome; LAMP1, lysosomes; NUP98, nuclear envelope; fibrillin, nucleolus and syntaxin 6, trans-Golgi network (data not shown). (f) Proximity-ligation assay (PLA) of FAMIN in combination with PMP70, Catalase (red) or centromere protein A (red) as negative control in THP-1 macrophages. DAPI, blue; scale bar = 10 μm. White box corresponds to enlarged images shown in Fig. 1e. Data are representative of three independent experiments.
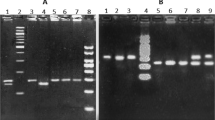
Supplementary Figure 3 Generation of mFamin–/–, mFaminp254I and mFaminp284R mice.
Schematic representation of the targeting strategy for creating a mFamin–/– allele in murine ES cells. The targeting vector, L1L2_Bact_P (International Mouse Knockout Consortium), contains a reading frame-independent LacZ gene trap cassette consisting of an En2 splice acceptor (SA), an internal ribosomal entry site (IRES), LacZ gene and a polyadenylation site (PA). The vector also contains a selectable marker consisting of a human β-actin promoter (hBactP), neomycin resistance gene (neo) and a PA site. The vector is flanked by flippase recognition target (FRT) sites to allow removal of the targeting cassette and conditional mFamin deletion upon Cre-mediated recombination of the loxP sites. mFaminp254I and mFaminp284R mice were generated by CRISPR/Cas9 genome editing to introduce nucleotide changes to encode p.254I or p.284R amino acids, respectively, at indicated positions. Nucleotides in mFamin exon 5 were targeted using guide RNAs, ‘line’ 7 and ‘line’ 9, respectively as outlined in the methods. Two different targeting oligodeoxynucleotides were used for each: one containing only the nucleotide changes leading to the amino acid substitutions, the other containing 2 additional synonymous nucleotide changes (‘wobble’) in the underlined region.
Supplementary Figure 4 Famin expression is highest in M1 macrophages, and FAMIN deficiency does not affect FASN expression.
(a) mRNA expression of Famin in M0, M1 and M2 macrophages (MΦ). (b) mRNA expression of Fasn in M0, M1 and M2 macrophages. (c) Immunoblots (IB) of FASN in M1 and M2 macrophage cell lysates; β-actin loading control. Data are from one experiment with three mice representative of two (b,c; mean ± S.E.M.) or three independent experiments (c).
Supplementary Figure 5 FAMIN does not directly affect the Krebs cycle.
(a) Schematic representation of 1,2-13C-glucose incorporation, via citrate, into fatty acyl species. (b) Basal extracellular acidification rate (ECAR) of mFamin–/– and mFamin+/+ M1 and M2 macrophages (MΦ) in the presence of exogenous pyruvate (n = 5/14). (c) Relative levels of malate, fumarate, succinate and α-ketoglutarate in M1 and M2 macrophages. (d) Oxygen consumption rate (OCR) of mFamin–/– and mFamin+/+ M2 macrophages treated as indicated with 40 μM etomoxir (ETO) 1 h prior to OCR measurement and followed by sequential treatment (dotted vertical lines) with oligomycin (Oligo), FCCP, and rotenone plus antimycin (Rot + ant). *P < 0.05, **P < 0.01 (Unpaired, two-tailed Student’s t-test). Data are pooled from three independent experiments (b; mean ± S.E.M.), from one experiment with seven mice per group (c; mean ± S.E.M.) or from one experiment with three mice representative of two independent experiments (d; mean ± S.E.M.).
Supplementary Figure 6 FAMIN-deficient M2 macrophages exhibit impaired mitochondrial ROS- and FAO-dependent production of extracellular ROS.
(a,b) Intracellular ROS measurement in unstimulated M1 and M2 mFamin–/– and mFamin+/+ macrophages (MΦ) stained with the cytosolic ROS indicator, 5-(and-6)-chloromethyl-2-7-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA) and measured in relative fluorescence units, RFU. (c–i) Zymosan induced eROS production in M1 and M2 macrophages treated as indicated for 16 h with 20 μM C75; or 1 h 40 μM etomoxir (ETO) or 500 μM mitoTEMPO prior to stimulation; or silenced for Fasn (Fasn siRNA), Cpt1a (Cpt siRNA) or Cybb (Cybb siRNA) or scrambled siRNA (Ctrl siRNA); Left, eROS kinetic plots measured in relative light units, RLU and right, area under curve, AUC. (j) NADPH quantification in M1 and M2 macrophages cell lysates. (k) PMA-induced eROS production in mFamin–/– and mFamin+/+ neutrophils. *P < 0.05, **P < 0.01 (Unpaired, two-tailed Student’s t-test). Data are from one experiment with three mice representative of two independent experiments (a–k; mean ± S.E.M.).
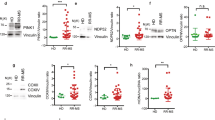
Supplementary Figure 7 FAMIN p.I254V and p.C284R M2 macrophage have impaired eROS production.
(a) Zymosan-stimulated eROS production in mFamin–/–, mFaminp254V, mFaminp254I and mFaminp284R murine M2 macrophages (MΦ). (b) Zymosan induced eROS production in mFaminp254I and mFaminp254V M2 macrophages treated as indicated for 16 h with 20 μM C75. (c) FAMIN mRNA expression in M2 macrophages and neutrophils from healthy donors homozygous for the Crohn’s disease and leprosy risk (‘rs3764147G/G’) and non-risk (‘rs3764147A/A’) haplotypes. Data are from one experiment with three mice representative of three independent experiments (a,b; mean ± S.E.M.) or pooled from ten independent experiments (c; mean ± S.E.M.).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Tables 1–5 (PDF 1676 kb)
Rights and permissions
About this article
Cite this article
Cader, M., Boroviak, K., Zhang, Q. et al. C13orf31 (FAMIN) is a central regulator of immunometabolic function. Nat Immunol 17, 1046–1056 (2016). https://doi.org/10.1038/ni.3532
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3532
This article is cited by
-
The 4th NextGen Therapies for SJIA and MAS: part 1 the elephant in the room: diagnostic/classification criteria for systemic juvenile idiopathic arthritis and adult-onset still’s disease
Pediatric Rheumatology (2024)
-
Effects of arginine on coenzyme-Q10 micelle uptake for mitochondria-targeted nanotherapy in phenylketonuria
Drug Delivery and Translational Research (2024)
-
Monogene Varianten in „laccase domain-containing 1“ (LACC1) als Ursache einer juvenilen Arthritis
Zeitschrift für Rheumatologie (2024)
-
Harnessing metabolism of hepatic macrophages to aid liver regeneration
Cell Death & Disease (2023)
-
Glucocorticoid receptor modulates myeloid-derived suppressor cell function via mitochondrial metabolism in immune thrombocytopenia
Cellular & Molecular Immunology (2022)