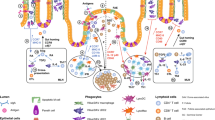
Abstract
Oral tolerance prevents pathological inflammatory responses to innocuous foreign antigens by peripheral regulatory T cells (pTreg cells). However, whether a particular subset of antigen-presenting cells (APCs) is required during dietary antigen exposure for the 'instruction' of naive CD4+ T cells to differentiate into pTreg cells has not been defined. Using myeloid lineage–specific APC depletion in mice, we found that monocyte-derived APCs were dispensable, while classical dendritic cells (cDCs) were critical, for pTreg cell induction and oral tolerance. CD11b− cDCs from the gut-draining lymph nodes efficiently induced pTreg cells and, conversely, loss of transcription factor IRF8–dependent CD11b− cDCs impaired their polarization, although oral tolerance remained intact. These data reveal the hierarchy of cDC subsets in the induction of pTreg cells and their redundancy during the development of oral tolerance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
References
Kretschmer, K. et al. Inducing and expanding regulatory T cell populations by foreign antigen. Nat. Immunol. 6, 1219–1227 (2005).
Mucida, D. et al. Oral tolerance in the absence of naturally occurring Tregs. J. Clin. Invest. 115, 1923–1933 (2005).
Curotto de Lafaille, M.A. et al. Adaptive Foxp3+ regulatory T cell-dependent and -independent control of allergic inflammation. Immunity 29, 114–126 (2008).
Josefowicz, S.Z. et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature 482, 395–399 (2012).
Ostroukhova, M. et al. Tolerance induced by inhaled antigen involves CD4+ T cells expressing membrane-bound TGF-beta and FOXP3. J. Clin. Invest. 114, 28–38 (2004).
Coombes, J.L. et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-β and retinoic acid-dependent mechanism. J. Exp. Med. 204, 1757–1764 (2007).
Mucida, D. et al. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science 317, 256–260 (2007).
Sun, C.M. et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med. 204, 1775–1785 (2007).
Hadis, U. et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity 34, 237–246 (2011).
Annacker, O. et al. Essential role for CD103 in the T cell-mediated regulation of experimental colitis. J. Exp. Med. 202, 1051–1061 (2005).
Mazzini, E., Massimiliano, L., Penna, G. & Rescigno, M. Oral tolerance can be established via gap junction transfer of fed antigens from CX3CR1+ macrophages to CD103+ dendritic cells. Immunity 40, 248–261 (2014).
Farache, J. et al. Luminal bacteria recruit CD103+ dendritic cells into the intestinal epithelium to sample bacterial antigens for presentation. Immunity 38, 581–595 (2013).
Diehl, G.E. et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX3CR1hi cells. Nature 494, 116–120 (2013).
Worbs, T. et al. Oral tolerance originates in the intestinal immune system and relies on antigen carriage by dendritic cells. J. Exp. Med. 203, 519–527 (2006).
Schulz, O. et al. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J. Exp. Med. 206, 3101–3114 (2009).
Iwata, M. et al. Retinoic acid imprints gut-homing specificity on T cells. Immunity 21, 527–538 (2004).
Johansson-Lindbom, B. et al. Functional specialization of gut CD103+ dendritic cells in the regulation of tissue-selective T cell homing. J. Exp. Med. 202, 1063–1073 (2005).
Idoyaga, J. et al. Specialized role of migratory dendritic cells in peripheral tolerance induction. J. Clin. Invest. 123, 844–854 (2013).
Yamazaki, S. et al. CD8+ CD205+ splenic dendritic cells are specialized to induce Foxp3+ regulatory T cells. J. Immunol. 181, 6923–6933 (2008).
Schreiber, H.A. et al. Intestinal monocytes and macrophages are required for T cell polarization in response to Citrobacter rodentium. J. Exp. Med. 210, 2025–2039 (2013).
Meredith, M.M. et al. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J. Exp. Med. 209, 1153–1165 (2012).
Jung, S. et al. In vivo depletion of CD11c+ dendritic cells abrogates priming of CD8+ T cells by exogenous cell-associated antigens. Immunity 17, 211–220 (2002).
Niess, J.H. et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 307, 254–258 (2005).
Watchmaker, P.B. et al. Comparative transcriptional and functional profiling defines conserved programs of intestinal DC differentiation in humans and mice. Nat. Immunol. 15, 98–108 (2014).
Miller, J.C. et al. Immunological Genome Consortium. Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 13, 888–899 (2012).
Vander Lugt, B. et al. Transcriptional programming of dendritic cells for enhanced MHC class II antigen presentation. Nat. Immunol. 15, 161–167 (2014).
Aliberti, J. et al. Essential role for ICSBP in the in vivo development of murine CD8α+ dendritic cells. Blood 101, 305–310 (2003).
Schiavoni, G. et al. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8α+ dendritic cells. J. Exp. Med. 196, 1415–1425 (2002).
Edelson, B.T. et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8α+ conventional dendritic cells. J. Exp. Med. 207, 823–836 (2010).
Hildner, K. et al. Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100 (2008).
Holtschke, T. et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell 87, 307–317 (1996).
Rivollier, A., He, J., Kole, A., Valatas, V. & Kelsall, B.L. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J. Exp. Med. 209, 139–155 (2012).
Schraml, B.U. et al. Genetic tracing via DNGR-1 expression history defines dendritic cells as a hematopoietic lineage. Cell 154, 843–858 (2013).
Loschko, J. et al. Absence of MHC class II on cDCs results in microbial-dependent intestinal inflammation. J. Exp. Med. (in the press).
Feng, J. et al. IFN regulatory factor 8 restricts the size of the marginal zone and follicular B cell pools. J. Immunol. 186, 1458–1466 (2011).
Cassani, B. et al. Gut-tropic T cells that express integrin α4β7 and CCR9 are required for induction of oral immune tolerance in mice. Gastroenterology 141, 2109–2118 (2011).
Goubier, A. et al. Plasmacytoid dendritic cells mediate oral tolerance. Immunity 29, 464–475 (2008).
Darrasse-Jèze, G. et al. Feedback control of regulatory T cell homeostasis by dendritic cells in vivo. J. Exp. Med. 206, 1853–1862 (2009).
Kinnebrew, M.A. et al. Interleukin 23 production by intestinal CD103+CD11b+ dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity 36, 276–287 (2012).
Zigmond, E. et al. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity 40, 720–733 (2014).
Cerovic, V. et al. Intestinal CD103− dendritic cells migrate in lymph and prime effector T cells. Mucosal Immunol. 6, 104–113 (2013).
Scott, C.L. et al. CCR2+CD103− intestinal dendritic cells develop from DC-committed precursors and induce interleukin-17 production by T cells. Mucosal Immunol. 8, 327–339 (2015).
Tamura, T. et al. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J. Immunol. 174, 2573–2581 (2005).
Yamamoto, M. et al. Shared and distinct functions of the transcription factors IRF4 and IRF8 in myeloid cell development. PLoS One 6, e25812 (2011).
Suzuki, S. et al. Critical roles of interferon regulatory factor 4 in CD11bhighCD8α− dendritic cell development. Proc. Natl. Acad. Sci. USA 101, 8981–8986 (2004).
Lewis, K.L. et al. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity 35, 780–791 (2011).
Persson, E.K. et al. IRF4 transcription-factor-dependent CD103+CD11b+ dendritic cells drive mucosal T helper 17 cell differentiation. Immunity 38, 958–969 (2013).
Satpathy, A.T. et al. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat. Immunol. 14, 937–948 (2013).
Tussiwand, R. et al. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity 42, 916–928 (2015).
Wang, Y. et al. Chylomicrons promote intestinal absorption and systemic dissemination of dietary antigen (ovalbumin) in mice. PLoS One 4, e8442 (2009).
Acknowledgements
We thank M. Amoury and L. Cohn for thoughts for the beginning of the project; M. Nussenzweig (The Rockefeller University) for Csf1rLsL-DTR, zDCDTR and zDCCre knock-in mice and conceptual input; S. Mazmanian (California Institute of Technology) for germ-free C57BL/6 mice; B. Reis and V. Pedicord for assistance in mouse-dissection experiments, critical discussions and help in preparing the manuscript; K. Velinzon and N. Thomas for assistance in cell sorting; The Rockefeller University Genomics Center; A. Rogoz, T. Rendon, S. Gonzalez and the Rockefeller University Comparative Bioscience Center for animal care and genotyping; and the New York University histology core for lung histological staining. Supported by the Swiss National Foundation (D.E.), The US National Institutes of Health (1R56AI119062 to D.M.) and the Leona M. and Harry B. Helmsley Charitable Trust (D.E. and D.M.).
Author information
Authors and Affiliations
Contributions
D.E. designed the study, performed the experiments (unless stated otherwise below) and wrote the manuscript; J.L. generated the zDCCre mice and backcrossed them onto the Irf8fl/fl background, carried out their hematopoietic characterization (Fig. 7) and substantially contributed to the design of experiments; M.L. and V.J. contributed substantially to the establishment of oral-tolerance protocols and to the oral-tolerance experiments; T.Y.O. performed the bioinformatics analysis of the RNA-seq experiments; and D.M. initiated, designed and supervised the study, performed experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
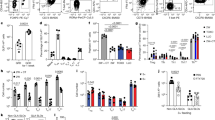
Supplementary Figure 1 Analysis of OVA uptake and clearance and APC depletion and replenishment in wild-type, MMDTR and zDCDTR chimeras.
(a) Lineage tree of major antigen presenting cell populations. Cells expressing zDC and depleted in zDCDTR mice are depicted in dark grey, cells expressing LysM and CSF1R and depleted in MMDTR mice are depicted in light grey. Macrophages can be monocyte– or yolk sack–derived and self-renewing (indicated by curved arrow). (b, c) Representative flow cytometry plots of small intestine lamina propria (LP), mLN (b), spleen, and blood (c) of WT, MMDTR and zDCDTR BMC mice 36h after DT injection. (d) Serum Flt3L levels in WT and zDCDTR BMC mice 24h post first DT injection. n=5, data representative of more than 20 experiments. (e) Coomassie-stained (top) and radiography of SDS-PAGE gel (bottom) loaded with indicated amount of 125I-labeled ovalbumin, showing purity and integrity of labeled protein. (f-h) Percentage of radioactivity recovered in the (f) urine and blood, (g) jejunum, inguinal lymph nodes, mLN and spleen and the (h) thyroid and liver of 8 weeks old C57BL/6 mice gavaged with 300,000 CPM 125I-OVA (4 μg) in 50 mg unlabeled OVA in 200 μl PBS over indicated time. n>2 per time-point. (i-m) Radioactivity in the (i) mLN, (j) spleen, (k) liver, (l) thyroid and (m) total blood of 16 weeks old WT, MMDTR and zDCDTR BMC mice 24 h after DT administration gavaged with 300,000 CPM 125I-OVA (4 μg) in 50 mg unlabeled OVA in 200 μl PBS and sacrificed 4 h later. n=3. (n) Analysis of total APC and subpopulation replenishment in the LP, Peyer’s patches (PP), mLN, spleen, and blood of WT, MMDTR and zDCDTR BMC mice 7 days post last DT injection. n=3. (o) Relative frequencies of TCRβ+CD4+, TCRβ+CD8+, and B220+ among CD45+ cells in WT, MMDTR and zDCDTR BMC mice 7 days post last DT injection. n=3. NS= not significant, *p<0.05, **p<0.01,***p<0.005.
Supplementary Figure 2 mLN cell counts and long-term pTreg cell induction in wild-type, MMDTR and zDCDTR chimeras.
(a) Total CD4+TCRβ+ cell numbers in the in the mLNs of WT, MMDTR and zDCDTR BMC mice, 60h post first DT treatment. (b) Percentage of CD45.1+ cells among CD4+TCRβ+ cells in the mLNs upon 2 doses of DT treatment without OVA gavage, analyzed 48h after first DT dose. (c) Percentage of CD45.1+ cells among CD4+TCRβ+ cells in the mLNs of WT, MMDTR and zDCDTR BMC mice, 7.5 days post first OVA gavage (see Fig. 3.a). (d) Cell division index and (e) total OT-II cell number in the mLNs of WT versus zDCDTR BMC mice, 7.5 days after first OVA gavage and 8 days post adoptive transfer of naïve OT-II cells. Each symbol indicates an individual mouse (d, e). n=4 per group, data (average±SEM) are representative of two independent experiments; NS= not significant, *p<0.05, **p<0.01,***p<0.005.
Supplementary Figure 3 Analysis of APC subset contributions to mLN APCs.
(a) Relative frequencies of APC subpopulations in the mLNs of eight weeks old GF versus SPF C57BL/6 mice. n=3. (b) Relative frequencies of APC subpopulations among CD45+ cells in the mLNs of GF versus SPF WT and zDCDTR BMC mice, 24h after DT administration. n=3. (c) Total CD45+ and total CD4+TCRβ+ cell counts in the mLNs of GF versus SPF WT and zDCDTR BMC mice, 24h after DT administration. n=3. (d) Percentage of CD45.1+ cells among total CD4+TCRβ+ cells in the mLNs of GF versus SPF WT and zDCDTR BMC mice, 24h after DT administration. n=3. (e) Total CD45+ and total CD4+TCRβ+ cell counts in the mLNs of CX3CR1LsL-DTR control (CTRL) and CX3CR1DTRBMC mice, 5 days after last DT administration. n=5 (f) Percentage of CD45.1+ cells among total CD4+TCRβ+ cells in the mLNs of CTRL and CX3CR1DTR BMC mice, 24h or 5 days after last DT administration. n=5. NS= not significant, *p<0.05, **p<0.01,***p<0.005.
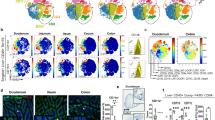
Supplementary Figure 4 cDC and lymphocyte frequencies in the mLNs draining the proximal-to-distal gastrointestinal tract.
(a) Dissection pictures of murine mLNs draining the duodenum (D), jejunum (J), ileum (I) and colon (C), indicated in white. Celiac (Ce) and pancreatic (P(d), adjacent to D, and P(j), adjacent to J) lymph nodes, the portal vein and the thoracic duct are indicated in black. Relative frequencies of (b) CD45+CD11c+, (c) CD11c+MHCIIint, (d) MHCIIintCD8α+CD11b−CD103+, (e) MHCIIintCD8α−CD11b+CD103−, (f) CD11c+MHCIIhi, (g) MHCIIhiCD11b−CD103+, (h) MHCIIhiCD11b+CD103+, (i) TCRβ−CD19+, (j) TCRβ+CD8+, (k) TCRβ+CD4+, (l) TCRβ+CD4+Foxp3+NRP+and (m) TCRβ+CD4+Foxp3+NRP− cells in the duodenum (D), jejunum (J), ileum (I) and colon (C) draining mLNs from 8 weeks old female C57BL/6 mice. Each symbol indicates an individual mouse (b-m). n=4. NS= not significant, *p<0.05, **p<0.01,***p<0.005. Data shown as average±SEM. (n) PCA of RNA-seq data from cDCs in the mLNs. Each circle represents average of triplicates per indicated cell population. (o-q) Heat map of (o) TGF-β synthesis, (p) retinoic acid synthesis and (q) cytokine gene cluster levels of DC subsets in the mLNs determined by RNA-seq. n=3.
Supplementary Figure 5 cDC subset depletion in the spleen of zDC(ΔIrf8) mice.
(a) Representative flow cytometry contour plots and (b) relative frequencies of CD8α+ and DEC205+ DCs and (c) total CD45+ counts in the spleen of Irf8fl/fl versus zDC(ΔIrf8) mice. n=4, data representative of more than six independent experiments. Each symbol indicates an individual mouse (b, c). NS= not significant, *p<0.05, **p<0.01,***p<0.005. Data shown as average±SEM.
Supplementary Figure 6 OT-II cell differentiation by IRF8 and IRF4-dependent cDCs from the mLNs.
PCA of RNA-seq data from OT-II cells co-cultured with indicated mLN cDC population. Each circle represents average of triplicates per cell population.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Table 1 (PDF 3715 kb)
Rights and permissions
About this article
Cite this article
Esterházy, D., Loschko, J., London, M. et al. Classical dendritic cells are required for dietary antigen–mediated induction of peripheral Treg cells and tolerance. Nat Immunol 17, 545–555 (2016). https://doi.org/10.1038/ni.3408
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3408
This article is cited by
-
Gamma-delta T cells modulate the microbiota and fecal micro-RNAs to maintain mucosal tolerance
Microbiome (2023)
-
Regulatory T cells in the face of the intestinal microbiota
Nature Reviews Immunology (2023)
-
Localization and movement of Tregs in gastrointestinal tract: a systematic review
Inflammation and Regeneration (2022)
-
Conventional type 1 dendritic cells protect against age-related adipose tissue dysfunction and obesity
Cellular & Molecular Immunology (2022)
-
Dendritic cell functions in the inductive and effector sites of intestinal immunity
Mucosal Immunology (2022)