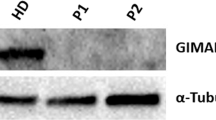
Abstract
The p110δ subunit of phosphatidylinositol-3-OH kinase (PI(3)K) is selectively expressed in leukocytes and is critical for lymphocyte biology. Here we report fourteen patients from seven families who were heterozygous for three different germline, gain-of-function mutations in PIK3CD (which encodes p110δ). These patients presented with sinopulmonary infections, lymphadenopathy, nodular lymphoid hyperplasia and viremia due to cytomegalovirus (CMV) and/or Epstein-Barr virus (EBV). Strikingly, they had a substantial deficiency in naive T cells but an over-representation of senescent effector T cells. In vitro, T cells from patients exhibited increased phosphorylation of the kinase Akt and hyperactivation of the metabolic checkpoint kinase mTOR, enhanced glucose uptake and terminal effector differentiation. Notably, treatment with rapamycin to inhibit mTOR activity in vivo partially restored the abundance of naive T cells, largely 'rescued' the in vitro T cell defects and improved the clinical course.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Okkenhaug, K. & Vanhaesebroeck, B. PI3K in lymphocyte development, differentiation and activation. Nat. Rev. Immunol. 3, 317–330 (2003).
Hemmings, B.A. & Restuccia, D.F. PI3K-PKB/Akt pathway. Cold Spring Harb. Perspect. Biol. 4, a011189 (2012).
Chantry, D. et al. p110δ, a novel phosphatidylinositol 3-kinase catalytic subunit that associates with p85 and is expressed predominantly in leukocytes. J. Biol. Chem. 272, 19236–19241 (1997).
Vanhaesebroeck, B. et al. P110δ, a novel phosphoinositide 3-kinase in leukocytes. Proc. Natl. Acad. Sci. USA 94, 4330–4335 (1997).
Huang, C.H. et al. The structure of a human p110α/p85α complex elucidates the effects of oncogenic PI3Kα mutations. Science 318, 1744–1748 (2007).
Gubser, P.M. et al. Rapid effector function of memory CD8 T cells requires an immediate-early glycolytic switch. Nat. Immunol. 14, 1064–1072 (2013).
Xu, X., Ye, L., Araki, K. & Ahmed, R. mTOR, linking metabolism and immunity. Semin. Immunol. 24, 429–435 (2012).
Yang, K. & Chi, H. mTOR and metabolic pathways in T cell quiescence and functional activation. Semin. Immunol. 24, 421–428 (2012).
Zinzalla, V., Stracka, D., Oppliger, W. & Hall, M.N. Activation of mTORC2 by association with the ribosome. Cell 144, 757–768 (2011).
Alessi, D.R. et al. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr. Biol. 7, 261–269 (1997).
Sarbassov, D.D., Guertin, D.A., Ali, S.M. & Sabatini, D.M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 (2005).
Kaech, S.M. & Cui, W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12, 749–761 (2012).
Finlay, D.K. et al. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J. Exp. Med. 209, 2441–2453 (2012).
Fox, C.J., Hammerman, P.S. & Thompson, C.B. Fuel feeds function: energy metabolism and the T-cell response. Nat. Rev. Immunol. 5, 844–852 (2005).
Astle, M.V. et al. AKT induces senescence in human cells via mTORC1 and p53 in the absence of DNA damage: implications for targeting mTOR during malignancy. Oncogene 31, 1949–1962 (2012).
Kim, E.H. & Suresh, M. Role of PI3K/Akt signaling in memory CD8 T cell differentiation. Front. Immunol. 4, 20 (2013).
Araki, K. et al. mTOR regulates memory CD8 T-cell differentiation. Nature 460, 108–112 (2009).
Kim, E.H. et al. Signal integration by Akt regulates CD8 T cell effector and memory differentiation. J. Immunol. 188, 4305–4314 (2012).
Sukumar, M. et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J. Clin. Invest. 123, 4479–4488 (2013).
Jarrett, A.F., Armstrong, A.A. & Alexander, E. Epidemiology of EBV and Hodgkin's lymphoma. Ann. Oncol. 7 (suppl. 4), 5–10 (1996).
Berndt, A. et al. The p110 d structure: mechanisms for selectivity and potency of new PI(3)K inhibitors. Nat. Chem. Biol. 6, 117–124 (2010).
Bader, A.G., Kang, S., Zhao, L. & Vogt, P.K. Oncogenic PI3K deregulates transcription and translation. Nat. Rev. Cancer 5, 921–929 (2005).
Mandelker, D. et al. A frequent kinase domain mutation that changes the interaction between PI3Kα and the membrane. Proc. Natl. Acad. Sci. USA 106, 16996–17001 (2009).
Wu, H. et al. Regulation of Class IA PI 3-kinases: C2 domain-iSH2 domain contacts inhibit p85/p110α and are disrupted in oncogenic p85 mutants. Proc. Natl. Acad. Sci. USA 106, 20258–20263 (2009).
Zhao, L. & Vogt, P.K. Helical domain and kinase domain mutations in p110α of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc. Natl. Acad. Sci. USA 105, 2652–2657 (2008).
Jou, S.T. et al. Identification of variations in the human phosphoinositide 3-kinase p110δ gene in children with primary B-cell immunodeficiency of unknown aetiology. Int. J. Immunogenet. 33, 361–369 (2006).
Damgaard, R.B. et al. Disease-causing mutations in the XIAP BIR2 domain impair NOD2-dependent immune signalling. EMBO Mol. Med. 5, 1278–1295 (2013).
Li, F.Y. et al. Second messenger role for Mg2+ revealed by human T-cell immunodeficiency. Nature 475, 471–476 (2011).
Brenchley, J.M. et al. Expression of CD57 defines replicative senescence and antigen-induced apoptotic death of CD8+ T cells. Blood 101, 2711–2720 (2003).
Roux, P.P. et al. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J. Biol. Chem. 282, 14056–14064 (2007).
Okkenhaug, K. et al. Impaired B and T cell antigen receptor signaling in p110δ PI 3-kinase mutant mice. Science 297, 1031–1034 (2002).
Fruman, D.A. et al. Impaired B cell development and proliferation in absence of phosphoinositide 3-kinase p85α. Science 283, 393–397 (1999).
Suzuki, H. et al. Xid-like immunodeficiency in mice with disruption of the p85alpha subunit of phosphoinositide 3-kinase. Science 283, 390–392 (1999).
Conley, M.E. et al. Agammaglobulinemia and absent B lineage cells in a patient lacking the p85alpha subunit of PI3K. J. Exp. Med. 209, 463–470 (2012).
Suzuki, A. et al. T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity 14, 523–534 (2001).
Denley, A., Kang, S., Karst, U. & Vogt, P.K. Oncogenic signaling of class I PI3K isoforms. Oncogene 27, 2561–2574 (2008).
Zhao, L. & Vogt, P.K. Class I PI3K in oncogenic cellular transformation. Oncogene 27, 5486–5496 (2008).
Vivanco, I. & Sawyers, C.L. The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat. Rev. Cancer 2, 489–501 (2002).
Billottet, C. et al. A selective inhibitor of the p110 d isoform of PI 3-kinase inhibits AML cell proliferation and survival and increases the cytotoxic effects of VP16. Oncogene 25, 6648–6659 (2006).
Sinclair, L.V. et al. Phosphatidylinositol-3-OH kinase and nutrient-sensing mTOR pathways control T lymphocyte trafficking. Nat. Immunol. 9, 513–521 (2008).
Omori, S.A. et al. Regulation of class-switch recombination and plasma cell differentiation by phosphatidylinositol 3-kinase signaling. Immunity 25, 545–557 (2006).
Srinivasan, L. et al. PI3 kinase signals BCR-dependent mature B cell survival. Cell 139, 573–586 (2009).
Amzel, L.M. et al. Structural comparisons of class I phosphoinositide 3-kinases. Nat. Rev. Cancer 8, 665–669 (2008).
Yentrapalli, R. et al. The PI3K/Akt/mTOR pathway is implicated in the premature senescence of primary human endothelial cells exposed to chronic radiation. PLoS ONE 8, e70024 (2013)
Gourley, T.S., Wherry, E.J., Masopust, D. & Ahmed, R. Generation and maintenance of immunological memory. Semin. Immunol. 16, 323–333 (2004).
Faivre, S., Kroemer, G. & Raymond, E. Current development of mTOR inhibitors as anticancer agents. Nat. Rev. Drug Discov. 5, 671–688 (2006).
Lannutti, B.J. et al. CAL-101, a p110δ selective phosphatidylinositol-3-kinase inhibitor for the treatment of B-cell malignancies, inhibits PI3K signaling and cellular viability. Blood 117, 591–594 (2011).
Angulo, I. et al. Phosphoinositide 3-kinase δ gene mutation predisposes to respiratory infection and airway damage. Science doi:10.1126/science.1243292 (17 October 2013).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Palendira, U. et al. Molecular pathogenesis of EBV susceptibility in XLP as revealed by analysis of female carriers with heterozygous expression of SAP. PLoS Biol. 9, e1001187 (2011).
Avery, D.T. et al. B cell-intrinsic signaling through IL-21 receptor and STAT3 is required for establishing long-lived antibody responses in humans. J. Exp. Med. 207, 155–171 (2010).
Acknowledgements
We thank the referring physicians, as well as the patients and families. Supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, Clinical Center of the US National Institutes of Health (C.L.L., H.S.K., J.E.N., M.B., J.S., W.O., V.K.R., A.A., A.A.H., K.N.O., T.A.F., S.P., S.M.H., J.I.C., M.J.L., G.U.), the National Human Genome Research Institute of the US National Institutes of Health (F.Z., J.L.C., P.L.S.), the Frederick National Laboratory for Cancer Research of the US National Institutes of Health (HHSN261200800001E), the National Health and Medical Research Council of Australia (E.K.D., U.P., S.G.T.), Cancer Council NSW (S.G.T.), the Cancer Institute NSW (U.P.), the Research Foundation-Flanders, Belgium (L.M.) and the National Institute of General Medical Sciences (C.L.L. and R.Z.). The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
Author information
Authors and Affiliations
Contributions
C.L.L did experiments, analyzed data and developed and wrote the manuscript; H.S.K., F.Z., E.K.D., U.P., D.T.A., L.M. and J.L.C. did experiments and analyzed data; J.E.N. analyzed genomic DNA sequencing and bioinformatics, discovered candidate genes and analyzed protein structure; M.B. analyzed p110 structure; J.S. did experiments and analyzed genomic DNA sequencing and data; W.O. did experiments; D.M.F. supervised research and data analysis; V.K.R. evaluated patients and collected data; T.P.A. and J.I.C. provided patient access, clinical data, samples and advice; A.A. evaluated patients and collected and analyzed data; A.A.H. coordinated patient access, data collection and analysis; L.R.F. evaluated and prepared data from clinical imaging studies; K.N.O. evaluated patients and collected and analyzed data; T.A.F. supervised research and data analysis and provided advice; S.P. did histological and immunohistochemical analyses of patient samples; S.M.H. supervised research and data analysis and provided advice; J.B.O planned and supervised whole-exome sequencing experiments; S.G.T. planned and supervised experiments, analyzed data, provided advice and prepared the manuscript; P.L.S. planned and supervised experiments, analyzed data and provided advice; M.J.L. supervised research and data analysis, provided advice and prepared the manuscript; G.U. coordinated research efforts, supervised research work and data analysis, and prepared the manuscript; and all authors discussed and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Note added in proof: Another study has now independently described a cohort of patients with the E1021K substitution of p110δ48.
Integrated supplementary information
Supplementary Figure 1 Lymphadenopathy, mitogen response, effector cells, and serum Ig assessment.
(a) Computerized Tomography (CT) images demonstrating lymphadenopathy (arrows) in patient F.II.1 and absence of spleen (removed surgically). (b) Three dimensional surface rendered image with cut plane at level of porta hepatis showing lymph node masses surrounding the portal vein and adjacent vasculature (darker red mass within white oval). (c) 3H-Thymidine incorporation in counts per minute (CPM) to assess proliferation of PBMCs after three days in response to the indicated stimulus. Each symbol represents an individual healthy control subject (Ctrl, n = 10) or patient (Pt, n = 10). Small horizontal lines indicate mean (± s.d.). *P < 0.0001, **P = 0.0002, ***P < 0.0001, ****P < 0.0001 (Mann-Whitney test). (d) Cumulative data for multiple healthy controls (Ctrl, n = 8) and patients (Pt, n = 9) showing % CCR7− negative cells among CD8+ T cells in the peripheral blood. Small horizontal lines indicate mean (± s.d.). *P < 0.0001 (unpaired t-test). (e) IgM (left) and IgA (right) serum concentrations as a function of the patient's age. Dotted lines indicate the upper and lower boundaries of the normal range from healthy subjects. Data are representative of ten independent experiments with one patient each (c), four independent experiments (d) or two to ten separate measurements over time for each of the nine indicated patients.
Supplementary Figure 2 Patient B cell defects.
(a) The proportion of total B cells (i.e., CD20+ cells) within the lymphocyte population of healthy controls (Ctrl, n = 6) compared to patients (Pt, n = 7). Small horizontal lines indicate mean (± s.d.). (b, c) Expression of (b) CD5 on transitional and naïve B cells and of (c) surface IgG and IgA on memory B cells from healthy controls (red histogram) and patients (blue histogram) was determined by flow cytometry. (d) CFSE-labeled naïve B cells were cultured with CD40L, CD40L+CpG, CD40L+anti-Ig or CD40L together with IL-4, IL-10, or IL-21 for 5 days before being assessed for dilution of the dye in healthy control (Ctrl, red) and patient (Pt, blue) samples. (e) Expression of AICDA mRNA was determined by quantitative PCR after stimulation of naïve B cells with the indicated stimulus. (f,g) Secretion of IgM, IgG, and IgA by stimulated naïve (f) and memory (g) B cells was determined by ELISA. Values for healthy controls (Ctrl) and patients (Pt) are given as mean ± sem of replicate cultures. *P = 0.03, **P = 0.005 (unpaired t-test). Data are representative of four independent experiments (b,c), two independent experiments (e), three independent experiments (f) or two independent experiments (g) or are from five independent experiments (a) or one experiment (d).
Supplementary Figure 3 Alignment of amino acid sequence of human p110α (PIK3CA) with that for human p110δ.
Residues mutated in our patients are shown in yellow, cyan and magenta.
Supplementary Figure 4 Hyperphosphorylation of Akt and intact association of mutant p110δ with p85α.
(a) Quantification of band intensities for the immunoblot shown in Fig. 2d in which serum-starved, activated T cells were assessed for p110δ, Akt, p-Akt (S473), and β-tubulin in patients D.II.1, D.II.2, and E.1 compared to three healthy controls (Ctrl) with 10 min anti-CD3 stimulation (+) or not (−). Total Akt values were normalized to β-tubulin abundance, then the ratio of p-Akt (S473) to normalized Akt was graphed. Small horizontal lines indicate mean (± s.d.). *P = 0.0002, **P = 0.01, ***P = 0.008 (unpaired t-test). (b) Summary of p-Akt (S473) by flow cytometry comparing mean fluorescence intensity (MFI). Basal p-Akt (S473) in paired (i.e., cultured for the same duration) T cell blasts from healthy controls (Ctrl, n = 5) and patients (Pt, n = 5) (left) or induced p-Akt (S473) after 10 min stimulation with anti-CD3 in T cell blasts from Ctrl (n = 3) and Pt (n = 3) (right, with mean (± s.d.)) are shown. *P = 0.02 (paired t-test), **P = 0.03 (unpaired t-test). (c) Immunoblots for the indicated proteins using PBMC lysates from patients with mutant p110δ or healthy controls (Ctrl) after stimulation with beads coated with anti-CD3 and anti-CD28 for 10 min (+) or not (?). (d,e) Representative histograms (left) and cumulative MFI data (right) for flow cytometry staining for p-Akt (T308) in patient (Pt, n = 3) versus healthy control (Ctrl, n = 3) T cell blasts, gating on CD4+ or CD8+ cells. Small horizontal lines indicate mean (± s.d.). *P = 0.03 (unpaired t-test). (f) Quantification of band intensities for the immunoblot shown in Fig. 2e together with data from two additional independent experiments. Numbers indicate the relative ratio (WT set to 1.0) of p-Akt (S473) to total Akt normalized for Flag expression. Small horizontal lines indicate mean (± s.d.). *P = 0.05, **P = 0.009 (unpaired t-test). (g) Immunoblot for p-Akt (S473) and β-tubulin on lysates from the human H9 T cell line overexpressing WT or the indicated mutant p110δ. (h) Immunoprecipitates (IP) of empty vector (EV), wild-type (WT), or mutant p110δ fused to a Flag epitope in HEK293T cells overexpressing p85α were blotted with antibodies against p85α, the Flag epitope, β-actin or immunoglobulin heavy chain (IgH). Data are from one experiment with three patients (a) or three independent experiments (f) or are representative of three independent experiments (b,d,e), four independent experiments with one patient each (c) or two independent experiments (g,h).
Supplementary Figure 5 Patient CD8+ T cells are hyperactivated with characteristics of enhanced effector function.
(a) Cumulative data for MHC class I-peptide tetramer (Tet) stains for EBV lytic (Ctrl n = 4 and Pt n = 4) and EBV latent antigens (Ctrl n = 3 and Pt n = 3). (b) Flow cytometry of gated EBV lytic and latent antigen-specific CD8+ T cells, stained for CCR7 and CD45RA. The percent of total events is shown in quadrants. (c) CD38 expression on the populations identified in (a) for patient G.1 (blue) compared to those for a healthy control (Ctrl, red). (d) Cumulative data for intracellular IFN-γ production in CD8+ T cell blasts from healthy controls (Ctrl, n = 3) versus patients (Pt, n = 4). Small horizontal lines indicate mean (± s.d.). P = 0.09 (unpaired t-test). (e) Cumulative data for granzyme B expression in CD4+ or CD8+ T cell blasts stimulated with low-dose, immobilized anti-CD3. Small horizontal lines indicate mean (± s.d.). *P = 0.003, **P = 0.01 (unpaired t-test). (f) Total LAMP-1 levels in activated CD8+ T cells from patients A.1 and G.1 compared to healthy control (Ctrl). (g) Cytolysis of P815 targets by anti-CD3-mediated redirected lysis for the indicated patients (Pt, n = 4) and healthy controls (Ctrl, n = 4). (h) Composite data for % CD57+ CD8+ T cells in patient (Pt, n = 4) versus healthy control (Ctrl, n = 4) PBMCs. Small horizontal lines indicate mean (± s.d.). *P = 0.002 (unpaired t-test). Data are from three (latent) or four (lytic) independent experiments (a) or three independent experiments (h) or are representative of four independent experiments (b,c,g) or two independent experiments (d,e,f).
Supplementary Figure 6 Expression of PD-1 on CD3+ cells in peripheral blood.
PBMCs were gated on CD3+ cells and assessed for CD45RA versus PD-1 expression in indicated patients (Pt, n = 7) and healthy controls (Ctrl, n = 2 adult and n = 2 pediatric). Data are representative of four independent experiments.
Supplementary Figure 7 Normal expression of signaling molecules in patient T cell blasts.
(a) Immunoblots for PTEN, PKC , p110δ, p-Erk, total Erk, and β-tubulin in T cell lysates from patients E.1 and F.II.1 or healthy control (Ctrl) cells stimulated with anti-CD3 for the times indicated in minutes (min). (b) Immunoblot for the PH domain-containing ARF6, p27kip1, and β-tubulin in indicated patient (n = 5 samples) and healthy control (Ctrl, n = 5 samples) T cell blasts after 10-min stimulation with anti-CD3 (+) or not (−). Data are from one experiment with two (a) or five (b) different patient samples.
Supplementary Figure 8 Increased, glucose- and amino acid-dependent basal S6 phosphorylation and glucose uptake in patient T cell blasts.
(a) Cumulative flow cytometric analysis of mean fluorescence intensity (MFI) for stimulated and unstimulated, permeabilized patient (Pt) or healthy control (Ctrl) T cell blasts after staining for p-S6 (S235, S236). Paired healthy control and patient blasts (i.e., cultured for the same duration) are connected by a line. *P = 0.04 (paired t-test). (b) Data from the same experiment as that shown in Fig. 4c but where cells were first rested in PBS for 2 hr before flow cytometric analysis of p- S6 (S235, S236) in unstimulated T cell blasts from patients G.1, D.II.1, and D.II.2 compared to healthy controls (Ctrl). (c) Glucose uptake by flow cytometric analysis of 2-NBDG fluorescence (linear MFI) after a 20-minute incubation period in glucose-starved T cell blasts from healthy controls (Ctrl, n = 3) and patients (Pt, n = 3). Paired healthy control and patient blasts (i.e., cultured for the same duration) are connected by a line. *P = 0.03 (paired t-test). Data are from three independent experiments (a) or one experiment with three different patients (b) or are representative of five independent experiments (c).
Supplementary Figure 9 CT images comparing hepatosplenomegaly and lymphadenopathy in patient A.1 before and after treatment with rapamycin.
Coronal reformations at the level of the tracheal bifurcation reveal more severe hepatosplenomegaly, hilar and mediastinal adenopathy, and splaying of tracheal bifurcation before (a) compared to after (b) rapamycin treatment.
Supplementary information
Supplementary Figures and Supplementary Tables
Supplementary Figures 1–9 and Supplementary Tables 1–3 (PDF 3795 kb)
Rights and permissions
About this article
Cite this article
Lucas, C., Kuehn, H., Zhao, F. et al. Dominant-activating germline mutations in the gene encoding the PI(3)K catalytic subunit p110δ result in T cell senescence and human immunodeficiency. Nat Immunol 15, 88–97 (2014). https://doi.org/10.1038/ni.2771
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2771
This article is cited by
-
Epstein–Barr virus-associated B-cell lymphoproliferative disorder meeting the definition of CAEBV B cell disease: a case report
BMC Infectious Diseases (2023)
-
Autoimmune lymphoproliferative immunodeficiencies (ALPID) in childhood: breakdown of immune homeostasis and immune dysregulation
Molecular and Cellular Pediatrics (2023)
-
Beyond PI3Ks: targeting phosphoinositide kinases in disease
Nature Reviews Drug Discovery (2023)
-
Activated Phosphoinositide 3-Kinase δ Syndrome: a Large Pediatric Cohort from a Single Center in China
Journal of Clinical Immunology (2022)
-
A variant in human AIOLOS impairs adaptive immunity by interfering with IKAROS
Nature Immunology (2021)