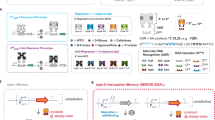
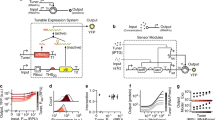
Abstract
Logic and memory are essential functions of circuits that generate complex, state-dependent responses. Here we describe a strategy for efficiently assembling synthetic genetic circuits that use recombinases to implement Boolean logic functions with stable DNA-encoded memory of events. Application of this strategy allowed us to create all 16 two-input Boolean logic functions in living Escherichia coli cells without requiring cascades comprising multiple logic gates. We demonstrate long-term maintenance of memory for at least 90 cell generations and the ability to interrogate the states of these synthetic devices with fluorescent reporters and PCR. Using this approach we created two-bit digital-to-analog converters, which should be useful in biotechnology applications for encoding multiple stable gene expression outputs using transient inputs of inducers. We envision that this integrated logic and memory system will enable the implementation of complex cellular state machines, behaviors and pathways for therapeutic, diagnostic and basic science applications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hasty, J., McMillen, D. & Collins, J.J. Engineered gene circuits. Nature 420, 224–230 (2002).
Regot, S. et al. Distributed biological computation with multicellular engineered networks. Nature 469, 207–211 (2011).
Auslander, S., Auslander, D., Muller, M., Wieland, M. & Fussenegger, M. Programmable single-cell mammalian biocomputers. Nature 487, 123–127 (2012).
Tamsir, A., Tabor, J.J. & Voigt, C.A. Robust multicellular computing using genetically encoded NOR gates and chemical 'wires'. Nature 469, 212–215 (2011).
Ajo-Franklin, C.M. et al. Rational design of memory in eukaryotic cells. Genes Dev. 21, 2271–2276 (2007).
Gardner, T.S., Cantor, C.R. & Collins, J.J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000).
Friedland, A.E. et al. Synthetic gene networks that count. Science 324, 1199–1202 (2009).
Bonnet, J., Subsoontorn, P. & Endy, D. Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc. Natl. Acad. Sci. USA 109, 8884–8889 (2012).
Benenson, Y. Biomolecular computing systems: principles, progress and potential. Nat. Rev. Genet. 13, 455–468 (2012).
Ham, T.S., Lee, S.K., Keasling, J.D. & Arkin, A.P. Design and construction of a double inversion recombination switch for heritable sequential genetic memory. PLoS ONE 3, e2815 (2008).
Ghosh, P., Pannunzio, N.R. & Hatfull, G.F. Synapsis in phage Bxb1 integration: selection mechanism for the correct pair of recombination sites. J. Mol. Biol. 349, 331–348 (2005).
Groth, A.C., Olivares, E.C., Thyagarajan, B. & Calos, M.P. A phage integrase directs efficient site-specific integration in human cells. Proc. Natl. Acad. Sci. USA 97, 5995–6000 (2000).
Callura, J.M., Cantor, C.R. & Collins, J.J. Genetic switchboard for synthetic biology applications. Proc. Natl. Acad. Sci. USA 109, 5850–5855 (2012).
Tabor, J.J. et al. A synthetic genetic edge detection program. Cell 137, 1272–1281 (2009).
Rinaudo, K. et al. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat. Biotechnol. 25, 795–801 (2007).
Gibson, D.G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Ringrose, L., Chabanis, S., Angrand, P.O., Woodroofe, C. & Stewart, A.F. Quantitative comparison of DNA looping in vitro and in vivo: chromatin increases effective DNA flexibility at short distances. EMBO J. 18, 6630–6641 (1999).
Cheng, A.A. & Lu, T.K. Synthetic biology: an emerging engineering discipline. Annu. Rev. Biomed. Eng. 14, 155–178 (2012).
Lu, T.K., Khalil, A.S. & Collins, J.J. Next-generation synthetic gene networks. Nat. Biotechnol. 27, 1139–1150 (2009).
Davis, J.H., Rubin, A.J. & Sauer, R.T. Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res. 39, 1131–1141 (2011).
Mijakovic, I., Petranovic, D. & Jensen, P.R. Tunable promoters in systems biology. Curr. Opin. Biotechnol. 16, 329–335 (2005).
Groth, A.C. & Calos, M.P. Phage integrases: biology and applications. J. Mol. Biol. 335, 667–678 (2004).
Gordley, R.M., Gersbach, C.A. & Barbas, C.F. III. Synthesis of programmable integrases. Proc. Natl. Acad. Sci. USA 106, 5053–5058 (2009).
Lux, M.W., Bramlett, B.W., Ball, D.A. & Peccoud, J. Genetic design automation: engineering fantasy or scientific renewal? Trends Biotechnol. 30, 120–126 (2012).
Lu, T.K. & Collins, J.J. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc. Natl. Acad. Sci. USA 106, 4629–4634 (2009).
Ortiz, M.E. & Endy, D. Engineered cell-cell communication via DNA messaging. J. Biol. Eng. 6, 16 (2012).
You, L., Cox, R.S., Weiss, R. & Arnold, F.H. Programmed population control by cell-cell communication and regulated killing. Nature 428, 868–871 (2004).
Bacchus, W. et al. Synthetic two-way communication between mammalian cells. Nat. Biotechnol. 30, 991–996 (2012).
McMillen, D., Kopell, N., Hasty, J. & Collins, J.J. Synchronizing genetic relaxation oscillators by intercell signaling. Proc. Natl. Acad. Sci. USA 99, 679–684 (2002).
Sambrook, J., Fritsch, E.F. & Maniatis, T. Molecular cloning: a laboratory manual. Cold Spring Laboratory Press 2 (1989).
Acknowledgements
The bxb1 gene was a generous gift from G.F. Hatfull (Department of Biological Sciences, University of Pittsburgh), and the riboregulator plasmids were donated by J.J. Collins (Biomedical Engineering, Boston University). The authors thank R. Danial and A.A. Cheng for careful comments on the manuscript. This work was supported by an Office of Naval Research Multidisciplinary University Research Initiative (MURI) grant and the Defense Advanced Research Projects Agency (DARPA).
Author information
Authors and Affiliations
Contributions
T.K.L. conceived of this study. P.S. and J.Y. implemented, constructed and performed all experiments. All authors analyzed the data, discussed results and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
P.S., J.Y. and T.K.L. have filed a provisional application with the US Patent and Trademark Office on this work.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Table 1 and Supplementary Data (PDF 6618 kb)
Rights and permissions
About this article
Cite this article
Siuti, P., Yazbek, J. & Lu, T. Synthetic circuits integrating logic and memory in living cells. Nat Biotechnol 31, 448–452 (2013). https://doi.org/10.1038/nbt.2510
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.2510
This article is cited by
-
Sentinel cells programmed to respond to environmental DNA including human sequences
Nature Chemical Biology (2024)
-
Engineering intelligent chassis cells via recombinase-based MEMORY circuits
Nature Communications (2024)
-
A hybrid transistor with transcriptionally controlled computation and plasticity
Nature Communications (2024)
-
Synthetic circuits based on split Cas9 to detect cellular events
Scientific Reports (2023)
-
Next generation synthetic memory via intercepting recombinase function
Nature Communications (2023)