Abstract
T helper 17 (TH17) cells are critically involved in host defence, inflammation, and autoimmunity1,2,3,4,5. Transforming growth factor β (TGFβ) is instrumental in TH17 cell differentiation by cooperating with interleukin-6 (refs 6, 7). Yet, the mechanism by which TGFβ enables TH17 cell differentiation remains elusive. Here we reveal that TGFβ enables TH17 cell differentiation by reversing SKI–SMAD4-mediated suppression of the expression of the retinoic acid receptor (RAR)-related orphan receptor γt (RORγt). We found that, unlike wild-type T cells, SMAD4-deficient T cells differentiate into TH17 cells in the absence of TGFβ signalling in a RORγt-dependent manner. Ectopic SMAD4 expression suppresses RORγt expression and TH17 cell differentiation of SMAD4-deficient T cells. However, TGFβ neutralizes SMAD4-mediated suppression without affecting SMAD4 binding to the Rorc locus. Proteomic analysis revealed that SMAD4 interacts with SKI, a transcriptional repressor that is degraded upon TGFβ stimulation. SKI controls histone acetylation and deacetylation of the Rorc locus and TH17 cell differentiation via SMAD4: ectopic SKI expression inhibits H3K9 acetylation of the Rorc locus, Rorc expression, and TH17 cell differentiation in a SMAD4-dependent manner. Therefore, TGFβ-induced disruption of SKI reverses SKI–SMAD4-mediated suppression of RORγt to enable TH17 cell differentiation. This study reveals a critical mechanism by which TGFβ controls TH17 cell differentiation and uncovers the SKI–SMAD4 axis as a potential therapeutic target for treating TH17-related diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Weaver, C. T., Hatton, R. D., Mangan, P. R. & Harrington, L. E. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu. Rev. Immunol. 25, 821–852 (2007)
Korn, T., Bettelli, E., Oukka, M. & Kuchroo, V. K. IL-17 and Th17 cells. Annu. Rev. Immunol. 27, 485–517 (2009)
Miossec, P., Korn, T. & Kuchroo, V. K. Interleukin-17 and type 17 helper T cells. N. Engl. J. Med. 361, 888–898 (2009)
Singh, R. P. et al. Th17 cells in inflammation and autoimmunity. Autoimmun. Rev. 13, 1174–1181 (2014)
Patel, D. D. & Kuchroo, V. K. Th17 cell pathway in human immunity: lessons from genetics and therapeutic interventions. Immunity 43, 1040–1051 (2015)
Bettelli, E. et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 (2006)
Veldhoen, M., Hocking, R. J., Atkins, C. J., Locksley, R. M. & Stockinger, B. TGFβ in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 24, 179–189 (2006)
Mangan, P. R. et al. Transforming growth factor-beta induces development of the TH17 lineage. Nature 441, 231–234 (2006)
Manel, N., Unutmaz, D. & Littman, D. R. The differentiation of human TH-17 cells requires transforming growth factor-β and induction of the nuclear receptor RORγt. Nat. Immunol. 9, 641–649 (2008)
Volpe, E. et al. A critical function for transforming growth factor-β, interleukin 23 and proinflammatory cytokines in driving and modulating human TH-17 responses. Nat. Immunol. 9, 650–657 (2008)
Gu, A. D. et al. A critical role for transcription factor Smad4 in T cell function that is independent of transforming growth factor β receptor signaling. Immunity 42, 68–79 (2015)
Yang, X. O. et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity 29, 44–56 (2008)
Lee, P. P. et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15, 763–774 (2001)
Chu, G. C., Dunn, N. R., Anderson, D. C., Oxburgh, L. & Robertson, E. J. Differential requirements for Smad4 in TGFβ-dependent patterning of the early mouse embryo. Development 131, 3501–3512 (2004)
Chytil, A., Magnuson, M. A., Wright, C. V. & Moses, H. L. Conditional inactivation of the TGF-β type II receptor using Cre:Lox. Genesis 32, 73–75 (2002)
Shapiro-Shelef, M., Lin, K. I., Savitsky, D., Liao, J. & Calame, K. Blimp-1 is required for maintenance of long-lived plasma cells in the bone marrow. J. Exp. Med. 202, 1471–1476 (2005)
Massagué, J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630 (2012)
Wang, T., Gu, S., Ronni, T., Du, Y. C. & Chen, X. In vivo dual-tagging proteomic approach in studying signaling pathways in immune response. J. Proteome Res. 4, 941–949 (2005)
Deheuninck, J. & Luo, K. Ski and SnoN, potent negative regulators of TGF-β signaling. Cell Res. 19, 47–57 (2009)
Colmenares, C. et al. Loss of the SKI proto-oncogene in individuals affected with 1p36 deletion syndrome is predicted by strain-dependent defects in Ski−/− mice. Nat. Genet. 30, 106–109 (2002)
Doyle, A. J. et al. Mutations in the TGF-β repressor SKI cause Shprintzen-Goldberg syndrome with aortic aneurysm. Nat. Genet. 44, 1249–1254 (2012)
Sun, Y., Liu, X., Ng-Eaton, E., Lodish, H. F. & Weinberg, R. A. SnoN and Ski protooncoproteins are rapidly degraded in response to transforming growth factor β signaling. Proc. Natl Acad. Sci. USA 96, 12442–12447 (1999)
Wu, J. W. et al. Structural mechanism of Smad4 recognition by the nuclear oncoprotein Ski: insights on Ski-mediated repression of TGF-β signaling. Cell 111, 357–367 (2002)
Nomura, T. et al. Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev. 13, 412–423 (1999)
Heldin, C. H., Miyazono, K. & ten Dijke, P. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature 390, 465–471 (1997)
Laurence, A. et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity 26, 371–381 (2007)
Yang, X. O. et al. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 282, 9358–9363 (2007)
Durant, L. et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity 32, 605–615 (2010)
Platt, R. J. et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014)
Labun, K., Montague, T. G., Gagnon, J. A., Thyme, S. B. & Valen, E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 44, W272–W276 (2016)
Acknowledgements
We thank E. Robertson and E. Bikoff for Smad4fl/fl mice, H. Moses for Tgfbr2fl/fl mice, D. Littman for Rorc−/− mice, F. Zhang for Cre-dependent Cas9 knock-in mice, N. Fisher for cell sorting, W. Chen and D. Zhang for discussion, and J. Massagué for the suggestion on SMAD4 ChIP–seq analysis. This study was supported by the National Natural Science Foundation of China (81402549, LJQ2015033) (G.Z.), the National Institutes of Health (NIH) (AI029564) and the National Multiple Sclerosis Society (CA10068) (J.P.Y.T.), the Intramural Research Program of the National Institute of Environmental Health Science (ES101965 to P.A.W. and ES102025 to D.N.C.), and by the NIH (AI097392; AI123193), the National Multiple Sclerosis Society (RG4654), and a Yang Family Biomedical Scholars Award (Y.Y.W.).
Author information
Authors and Affiliations
Contributions
S.Z. contributed to the design and implementation of the cellular, molecular, biochemical, and animal experiments, and the writing of the manuscript. M.T., X.X., S.Y.T., P.A.W., and D.N.C. contributed to ChIP–seq and RNA-seq experiments and bio-informatic analysis. L.Z., Q.K., and X.C. contributed to proteomic and biochemical experiments and data analysis. A.D.G. contributed to the in vitro assays. W.C. and J.P.T. contributed to the EAE experiments. G.Z. contributed to ChIP analysis. B.W. contributed to qRT–PCR analysis. J.S.S. contributed critical reagents. Y.Y.W. conceived the project, designed experiments, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks T. Egawa and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 TH17 cell differentiation in the absence of SMAD4.
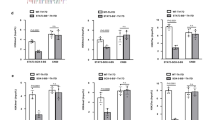
a, Naive CD4+ T cells isolated from wild-type and Cd4-cre;Smad4fl/fl (S4 KO) mice were activated in the presence of TGFβR inhibitor (i), IL-6 + TGFβR inhibitor (IL-6 + inhibitor), or IL-6 + TGFβ (IL-6 + TGFβ). IL-17A+ cells were assessed by flow cytometry 1 and 2 days later. Representative results (left) and statistical analysis (right) of five experiments are shown. b, The percentages of IL-17A+CD4+ and IFN-γ+CD4+ cells in the peripheral lymph nodes (pLN) and spleens from wild-type, Cd4-cre;Smad4fl/fl (S4 KO), Cd4-cre;Tgfbr2fl/fl (RII KO), and Cd4-cre;Smad4fl/fl;Tgfbr2fl/fl (S4–RII DKO) mice under steady state were assessed by flow cytometry. Representative results (left) and statistics from eight mice (right) are shown. (***P < 0.001; two-sided t-test; NS, not significant; centres indicate mean values.)
Extended Data Figure 2 SMAD4 suppresses RORγt expression.
a, CD4+ T cells from wild-type and Cd4-cre;Smad4fl/fl (S4 KO) mice were activated in the presence of IL-6 and TGFβR inhibitor. The mRNA expression of TH17-related genes was analysed at the indicated time points after activation by qRT–PCR. Means ± s.d. of three experiments are shown. b, Naive CD4+ T cells from wild-type and Cd4-cre;Smad4fl/fl (S4 KO) mice were sorted and activated in the presence of IL-6 and TGFβR inhibitor for 3 and 12 h. Total RNA was then collected for RNA-seq analysis. All genes were analysed and presented as volcano plots based on the log2(fold change) of SMAD4 knockout versus wild type and −log10(P value). Differentially expressed genes (P < 0.05) are highlighted in red. c, Naive CD4+ T cells from wild-type and Cd4-cre;Smad4fl/fl (S4 KO) mice were sorted and activated in the presence of IL-6 and TGFβR inhibitor. The mRNA expression of TH17-related genes was analysed at the indicated time points after activation by qRT–PCR. Means ± s.d. of three experiments are shown. d, Naive CD4+ T cells from wild-type and Cd4-cre;Smad4fl/fl;Tgfbr2fl/fl (S4–RII DKO) mice were sorted and activated in the presence of IL-6. The mRNA expression of TH17-related genes was analysed at the indicated time points after activation by qRT–PCR. Means ± s.d. of three experiments are shown. e, Naive CD4+ T cells from wild-type and Cd4-cre;Smad4fl/fl (S4 KO) mice were sorted and activated in the presence of IL-6 and TGFβR inhibitor. The RORγt protein expression was assessed by immunoblotting 1 and 4 days after activation. Results are representative of three experiments with similar results. f, CD4+ T cells from wild-type and Cd4-cre;Tgfbr2fl/fl;Smad4fl/fl (S4–RII DKO) mice were activated in the presence of IL-6. The RORγt protein expression was assessed by immunoblotting 1 day after activation. Results are representative of three experiments with similar results. g, CD4+ T cells from wild-type and Cd4-cre;Smad4fl/fl (S4 KO) mice were activated in the presence of IL-6 and TGFβR inhibitor. Cells were harvested after 12 h. ChIP assay was performed with control IgG antibody and SMAD4 antibody. The enrichment of SMAD4 binding to the Rorc promoter was determined. Means ± s.d. of three samples in one experiment of three are shown. h, CD4+ T cells from wild-type and Cd4-cre;Smad4fl/fl (S4 KO) mice were activated in the presence of IL-6 and TGFβR inhibitor. Cells were harvested after 12 h. ChIP–seq assay was performed with SMAD4 antibody. The enrichment of SMAD4 binding to the Il17a and Il17f loci was determined by the mapped read coverage of SMAD4 ChIP–seq data. The results of two independent experiments were show as ‘#1’ and ‘#2’. (*P < 0.05, **P < 0.01, ***P < 0.001; two-sided t-test; NS, not significant).
Extended Data Figure 3 SKI identification and its degradation upon low doses of TGFβ.
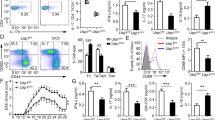
a, Schematic of quantitative immunoprecipitation and mass spectrometry proteomic strategy to identify SMAD4-binding proteins under different conditions. In one scheme, to identify SMAD4-binding protein in the absence of TGFβ signalling, CD4+ T cells from Cd4-cre;Smad4fl/fl (S4 KO) mice were sorted and activated in the presence of IL-6 + inhibitor in the SILAC/AACT medium provided either with amino acids containing light (L) isotopes or with amino acids containing heavy (H) isotopes. Cells were then transduced with retroviruses expressing either Flag tag (RV Flag) or Flag tag and SMAD4 fusion protein (RV Flag-S4). Cells were harvested and mixed 4 days after activation. Immunoprecipitation was performed using anti-Flag. Immunoprecipitated proteins were processed and subjected to quantitative mass spectromtery analysis. Proteins with a heavy/light ratio of more than 2 were identified. In the other scheme, to identify the proteins whose SMAD4 interaction was perturbed upon TGFβ stimulation, CD4+ T cells from wild-type mice were sorted and activated either in the presence of IL-6 + inhibitor in the SILAC/AACT medium provided with amino acids containing light isotopes or in the presence of IL-6 + TGFβ in the SILAC/AACT medium provided with amino acids containing heavy isotopes. Cells were harvested and mixed 4 days after activation. Immunoprecipitation was performed using anti-SMAD4 antibody. Immunoprecipitated proteins were processed and subjected to quantitative mass spectrometry analysis. Proteins with a heavy/light ratio of less than 1 were identified. The commonly identified protein SKI in the two experiments was subjected to further investigation. b, CD4+ T cells from wild-type mice were activated in the presence of IL-6 and the indicated doses of TGFβ. Cells were harvested after 24 h. SKI protein expression was detected by immunoblotting. Results are representative of three experiments with similar results. c, CD4+ T cells from wild-type mice were activated in the presence of IL-6 and the indicated doses of TGFβ. IL-17A+ and Foxp3+ cells were assessed by flow cytometry on day 4. Representative results (top) and statistical analysis (bottom) of five experiments are shown (centres indicate mean values). d, CD4+ T cells from wild-type or Cd4-cre;Smad2fl/fl (S2 KO) mice were activated in the presence of IL-6 for 24 h. Cells were then stimulated with the indicated conditions for an additional 1 h with or without 10 μM SIS3 (specific inhibitor of Smad3 phosphorylation). SKI protein expression and Smad3 phosphorylation were assessed by immunoblotting. Results are representative of three experiments with similar results.
Extended Data Figure 4 SKI and SMAD4 cooperatively suppress TH17 cell differentiation.
a, Bone marrow cells were isolated from the femur bones of sex- and age-matched Cd4-cre;CdC (Cas9, CD45.2+) mice and wild-type (wild type, CD45.1+) mice. Cells were mixed, and transduced with two different gRNA-expressing viruses (as indicated) and transferred into sublethally irradiated (400 cGy) Rag1−/− recipient mice. CD4+ T cells isolated from lymph nodes and spleen of generated bone marrow chimaeric mice were activated in the presence of IL-6 and TGFβR inhibitor. Cells transduced with gRNA in wild-type donors are indicated as wild type. Cells transduced with gRNA in CD4-cre;CdC donor are indicated as Ski knockout. IL-17A+ cells were assessed by flow cytometry on day 4. Representative results (left) and statistical analysis (right) of five experiments are shown. b, CD4+ T cells from wild-type mice were activated in the presence of IL-6 and TGFβ, and then transduced with MSCV-IRES–GFP (RV), MSCV-SKI-IRES–GFP (RV SKI), or co-transduced with MSCV-SKI-IRES–GFP and MSCV-RORγt-IRES-Thy1.1 (RV SKI+RORγt) retroviruses. IL-17A expression of transduced (GFP+) or co-transduced (GFP+Thy1.1+) T cells was assessed by flow cytometry. Representative results (left) and statistical analysis (right) of five experiments are shown. c, CD4+ T cells from wild-type and Cd4-cre;Smad4fl/fl (S4 KO) mice were activated in the presence of IL-6 + TGFβR inhibitor (i) or IL-6 + TGFβ. Cells were harvested 3 days later. ChIP assay was performed with control IgG antibody or SKI antibody. The relative enrichment of SKI binding to the Rorc locus was determined. Means ± s.d. of three samples in one experiment of three are shown. (**P < 0.01, ***P < 0.001; two-sided t-test; NS, not significant; centres indicate mean values.)
Extended Data Figure 5 TGFβ superfamily signalling overcomes SKI–SMAD4 complex-mediated suppression of RORγt expression in activated T cells to enable TH17 cell differentiation.
a, RORγt expression is potentiated by IL-6–STAT3 signalling but restrained by the histone deacetylase activity-containing SKI–SMAD4 complex that associates with and deacetylates the Rorc locus. b, Additional TGFβ or activin signalling triggers SKI degradation. The disruption of SKI–SMAD4 complex dissociates histone deacetylase activity from the Rorc locus and enables RORγt expression and TH17 cell differentiation.
Supplementary information
Supplementary Figures
This file contains Supplementary Figure 1, the uncropped gels with size marker indications and Supplementary Figure 2, gating strategies for flow cytometry analysis.
Rights and permissions
About this article
Cite this article
Zhang, S., Takaku, M., Zou, L. et al. Reversing SKI–SMAD4-mediated suppression is essential for TH17 cell differentiation. Nature 551, 105–109 (2017). https://doi.org/10.1038/nature24283
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature24283
This article is cited by
-
Context-dependent TGFβ family signalling in cell fate regulation
Nature Reviews Molecular Cell Biology (2023)
-
SENP2 restrains the generation of pathogenic Th17 cells in mouse models of colitis
Communications Biology (2023)
-
TGFβ control of immune responses in cancer: a holistic immuno-oncology perspective
Nature Reviews Immunology (2023)
-
Intricacies of TGF-β signaling in Treg and Th17 cell biology
Cellular & Molecular Immunology (2023)
-
A transcription factor DAF-5 functions in Haemonchus contortus development
Parasites & Vectors (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.